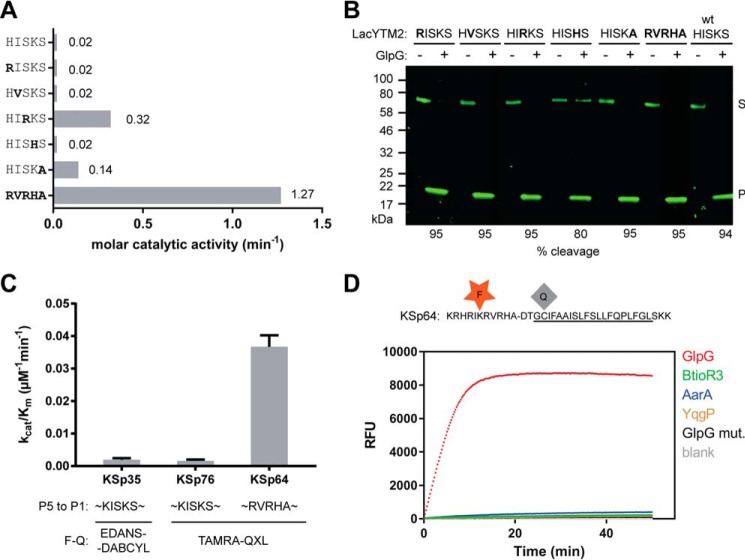
FIGURE 6.
The effect of non-prime side substitutions on the catalytic parameters and selectivity of rhomboid substrates. A, preferred amino acids in the P5 to P1 positions of the LacYTM2 transmembrane substrate improve its cleavage by GlpG. The LacYTM2 embedded in the MBP-thioredoxin chimera (18) was point-mutated in the P5 to P1 positions according to the sequence preferences of E. coli GlpG (19). The recombinant substrates were expressed in E. coli ΔglpG, purified, and molar catalytic activity of GlpG in cleaving each of the substrates was determined using gel-based assay (see “Experimental Procedures” for details). The concentration of substrate was always 1.47 μm, concentration of DDM was 0.5%(w/v), the concentration of GlpG was 0.8 μm for wild type substrate (HISKS). and for the RISKS, HVSKS, and HISHS mutants the concentration was 0.08 μm for the HISKA mutant and 0.016 μm for the HIRKS and RVRHA variants (to ensure reliable measurement of the initial reaction rate). Representative values from one of three independent experiments are shown. B, the effects of the preferred amino acids in the P5 to P1 region of LacYTM2 on the steady-state level of cleavage by GlpG in biological membranes in vivo. Plasmids encoding individual mutant versions of the chimeric mutant LacYTM2 substrates described above were transformed into E. coli MC4100 expressing endogenous GlpG, and 2 h after induction of expression of the substrates, the cell lysates were analyzed by immunoblotting using antibody against His tag, located at the C terminus of the constructs. Detection by near-infrared laser scanning, exhibiting linearity over 6 orders of magnitude, enabled reliable quantitation. Integration of product and substrate band intensities yielded steady-state substrate conversion values that are listed below the image. A representative experiment is displayed. C, apparent kinetic parameters of fluorogenic rhomboid substrates derived from LacYTM2. Initial reaction rates at very low substrate concentrations were used to calculate catalytic efficiency values (kcat/Km) of substrates KSp35, KSp64, and Ksp76 cleaved by GlpG at 0.5% (w/v) DDM. The reaction buffer was 20 mm HEPES, pH 7.4, 150 mm NaCl, 10% (v/v) DMSO, enzyme concentration was 0.4 μm, and substrate concentration ranged from 0.5 to 20 μm. Note that a mere optimization of the P5 to P1 region of the substrate increases the catalytic efficiency (kcat/Km) of its cleavage by GlpG by 23-fold. D, influence of the optimization of the P5 to P1 region on the selectivity of a transmembrane substrate for rhomboids. KSp76 underwent cleavage by rhomboid proteases GlpG, AarA, YqgP, and BtioR3 at the same concentrations as described in the legends to Figs. 3G and 5A. Note that optimization of the P5 to P1 region of the substrate increases the selectivity for GlpG dramatically.