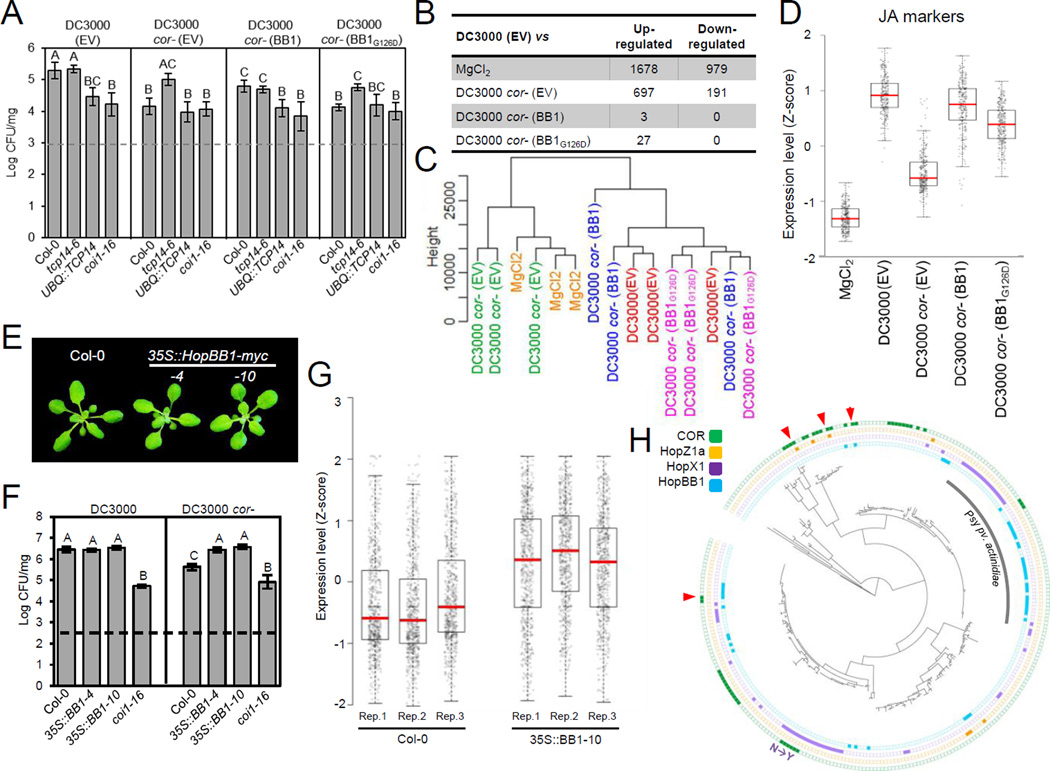
Figure 3. HopBB1 promotes bacteria growth and activates JA response.
(A) Bacterial-delivered HopBB1 promotes the growth of DC3000 cor- in Col-0. Two-week-old plants were spray inoculated with a bacteria suspension at OD600=0.2. CFU: Bacterial colony formation units. Error bars represent ±SD.
(B) Summary of transcriptional changes in Col-0 plants 24h after the treatment with DC3000 (EV), MgCl2 and the coronatine-deficient mutant strains carrying the empty vector (EV), HopBB1 or HopBB1G126D. Numbers represent the differentially expressed genes relative to the DC3000 treatment.
(C) Dendrogram constructed based on the entire transcriptome showing that the transcriptional signature of DC3000 cor- (EV), treated plants resembles that of the mock treatment; DC3000 cor- expressing either HopBB1 or HopBB1G126D trigger similar transcriptional responses as DC3000 (EV).
(D) A set of 253 JA marker genes are activated by DC3000 (EV) and/or DC3000 cor-(HopBB1); HopBB1G126D has reduced ability to activate these genes.
(E) Transgenic Arabidopsis plants expressing HopBB1 are morphologically indistinguishable from Col-0 wild-type. Bar=5mm
(F) Plants expressing HopBB1 complement the growth defects of DC3000 cor-.
(G) JA-responsive genes are activated in transgenic plants expressing HopBB1-myc. The z-score transformed expression of 672 JA responsive marker genes is shown for three biological replicates of Col-0 and transgenic plants expressing HopBB1-myc.
(H) The distribution of HopBB1, HopX1, HopZ1a and coronatine biosynthesis pathway in 287 sequenced Pseudomonas syringae genomes. Arrowhead: genomes contain two JA-activating tools. N→Y: A polymorphism (N→Y) exists in the HopX1 allele. See also Figure S3 and Tables S3 and S4.