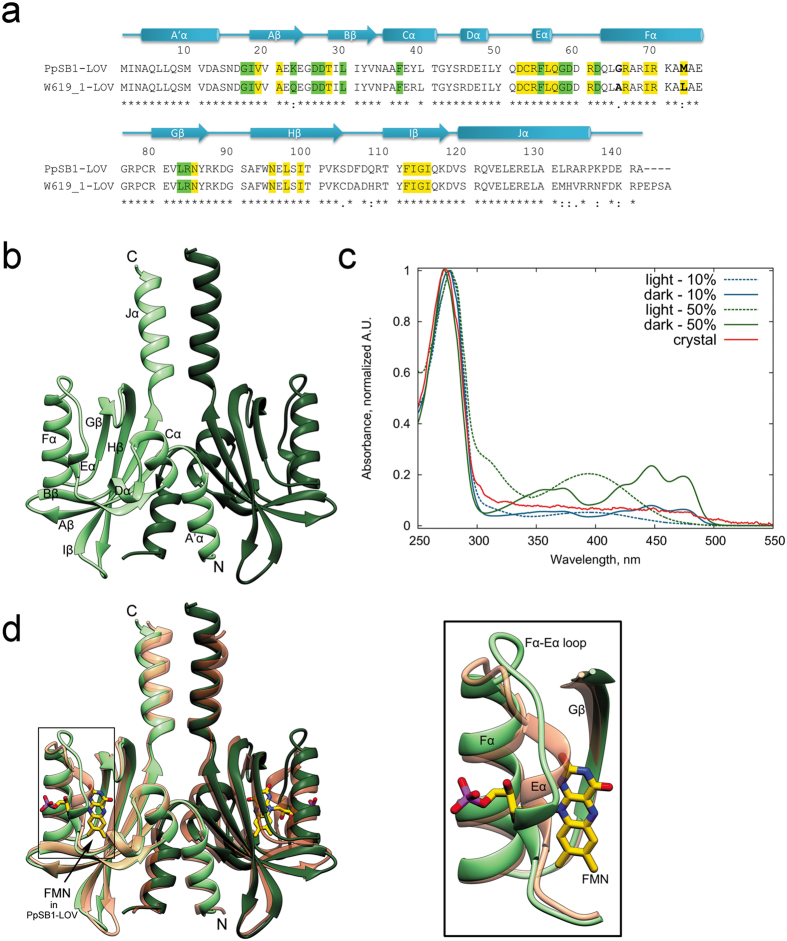
Figure 1. Apo-state crystal structure of W619_1-LOV.
(a) Sequence and secondary structure alignment of W619_1-LOV and PpSB1-LOV. Secondary structural elements (W619_1-LOV) and residue numbers are indicated above the sequence, where the beta strands from Aβ to Iβ are shown as arrows, and the helices from A’α to Jα are shown as cylinders. The asterisks at the bottom line of the alignment indicate identical residues at a given sequence position, while single and double dots refer to highly and moderately conserved residues, respectively. Residues lining the chromophore pocket at cut-off distances <4 Å and ≥4 Å from FMN in the PpSB1 crystal structure (PDB ID 5J3W) are highlighted in yellow and green, respectively. In the Fα helix, residues showing differences in sequence at position 65 and 73 are indicated in bold. (b) Ribbon representation of the W619_1-LOV dimer present in the asymmetric unit. Chains A and B are colored in light and dark shades. (c) Absorbance spectra of W619_1-LOV. Single crystal microspectrometry UV/Vis spectrum of a W619_1-LOV crystal shown as solid line in red color does not show any absorbance peaks that are typical for protein-bound flavin chromophores. In contrast, spectra of 10% (blue) and 50% (green) loaded protein solution show the typical absorbance maxima of 390 nm and 447 nm in light (dashed) and dark (solid lines) state, respectively. Spectra are normalized at 280 nm. (d) Superposition of W619_1-LOV (green) and PpSB1-LOV dark state (salmon, PDB ID 5J3W) crystal structures. The inset shows conformational differences in the secondary structure elements lining the chromophore pocket such as elongation of Fα helix, shortening of the Eα-Fα loop and partial unfolding of the Eα helix. The chromophore FMN bound to PpSB1-LOV is depicted as stick model and is colored by element: carbon, yellow; nitrogen, blue; oxygen, red; phosphorus, purple. Chains A and B are colored in light and dark shades.