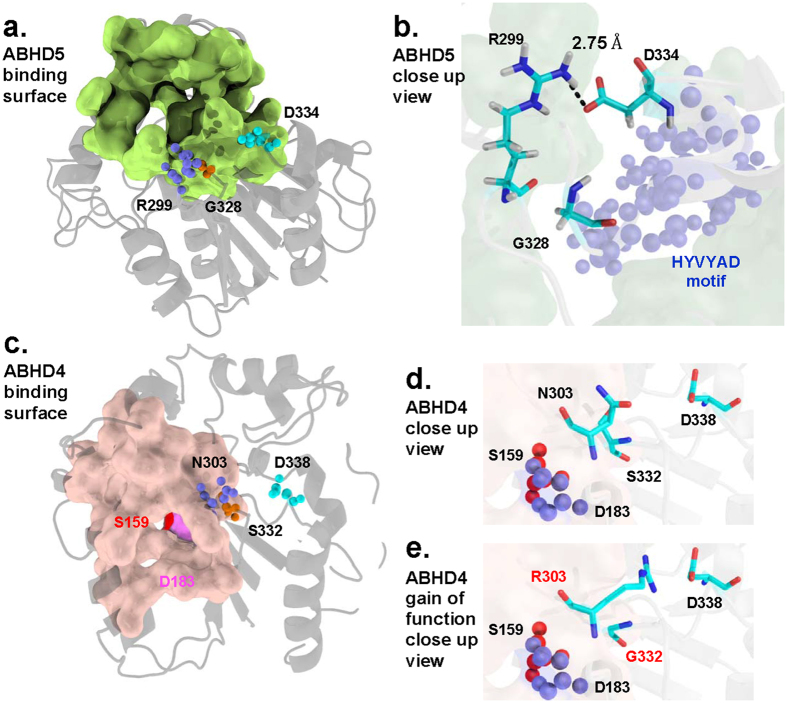
Figure 6. Shape analysis of ABHD5 and ABHD4 binding surfaces.
(a) The functionally important residues D334 (cyan), R299 (slate), and G328 (orange) in spheres are spatially clustered on the predicted ABHD5 binding surface (light green). (b) A close up view shows that residues G328 and D334 are centered around R299. The model predicts the local structure of ABHD5 may be stabilized by a salt-bridge between the D334 carboxyl group (red) and the R299 guanidinium group (blue). The HYVYAD motif containing D334 is shown in blue spheres. (c) The predicted catalytic ABHD4 residues D183 (violet) and nucleophile S159 (red) form a dyad located inside the deep tunnel of the identified ABHD4 binding surface (light orange). ABHD4 N303 (slate), S332 (orange) and D338 (cyan) are spatially oriented similarly to the corresponding key residues R299, G328, and D334 in ABHD5 (a) and are situated away from the ABHD4 catalytic center S159. (d) Close up view of ABHD4 N303, S332, and D338. Note that the region occupied by N303, S332 and D338 is spatially remote from the ABHD4 catalytic residues S159 and D183 (spheres). (e) In ABHD4 N303R/S332G, R303, G332 and D338 are spatially oriented similarly to R299, G328, and D334 in ABHD5 (b), conferring on ABHD4 N303R/S332G the ability to activate ATGL.