Figure 1. Analysis by molecular hybridization for detecting PLMVd in commercial peach samples.
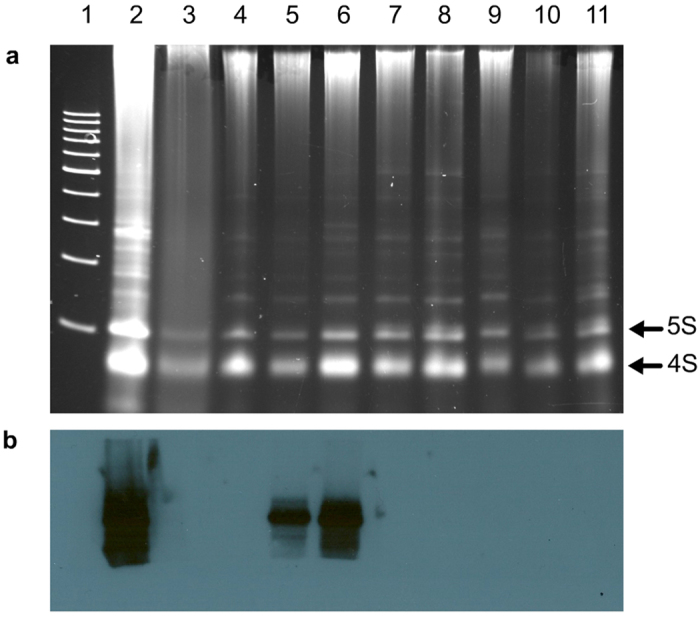
(a) Non-denaturing electrophoresis in a 5% polyacrylamide gel stained with ethidium bromide. Arrows on the right indicate the bands generated by 5S and 4S RNAs. (b) RNA gel-blot hybridization, with a full-length digoxigenin-labeled riboprobe for detecting PLMVd (+) strands, of the RNAs migrating in gel segment delimited by the 200- and 500-bp DNA size markers. Lane 1, 100-bp DNA multimers size markers. Lanes 2 and 3, RNA preparations from PLMVd-infected and mock-inoculated GF305 peach seedlings, respectively. Lanes 4 to 11, RNA preparations from commercial peach trees. The intense hybridization signals observed in the positive control (lane 2) and in two of the samples (V1 and V2, lanes 5 and 6, respectively) correspond to a size around the 300-bp marker. Even if the RNA amount loaded in the mock-inoculated control (lane 3) was lower than in some samples (e.g. lane 6), it was similar to another one that generated a strong hybridization signal (lane 5).
