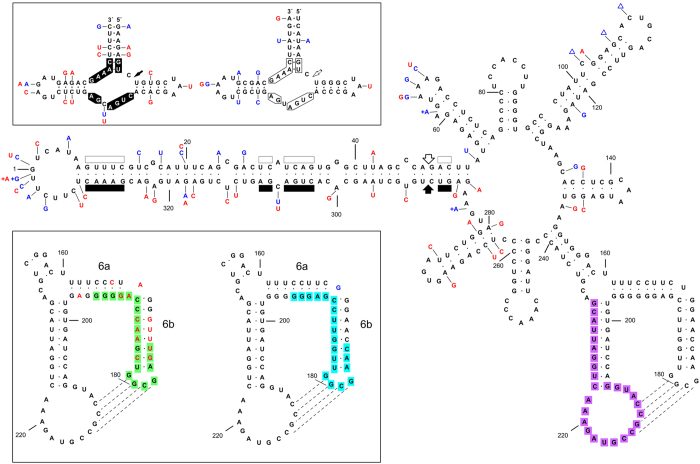
Figure 2. Primary and proposed secondary structure for the PLMVd plus strand of the reference variant GenBank M83545. 18 with two minor corrections10.
Changes in the representative symptomatic variant of class I (gds6)10 and in the representative variant of class II (v1.1) (this work), are denoted with blue and red characters, respectively. Symbols (+) and (Δ) refer to insertions and deletions, respectively, and broken lines to the kisssing-loop interaction39. Upper inset, hammerhead structures of the PLMVd plus and minus strands with the self-cleavage sites marked with arrows. Substitutions in variants gds6 and v1.1 do not disrupt the helices flanking the central core of 13 nucleotide residues (boxed) conserved in most natural hammerhead structures of viroid and viroid-like satellite RNAs. The same numbering is used for both polarities. Lower inset, domain 6a/6b harboring characteristic changes between variants of both classes. Note that helix 6a of variant v1.1 is one base-pair shorter. Fragments covered by the TaqMan rtRT-PCR probes recognizing variants of class I, class II, and of both classes, are indicated with green, blue and purple backgrounds, respectively. Sequence changes in variants gds6 and v1.1 may result in minor rearrangements of the secondary structure not represented here (see Supplementary Fig. S1).