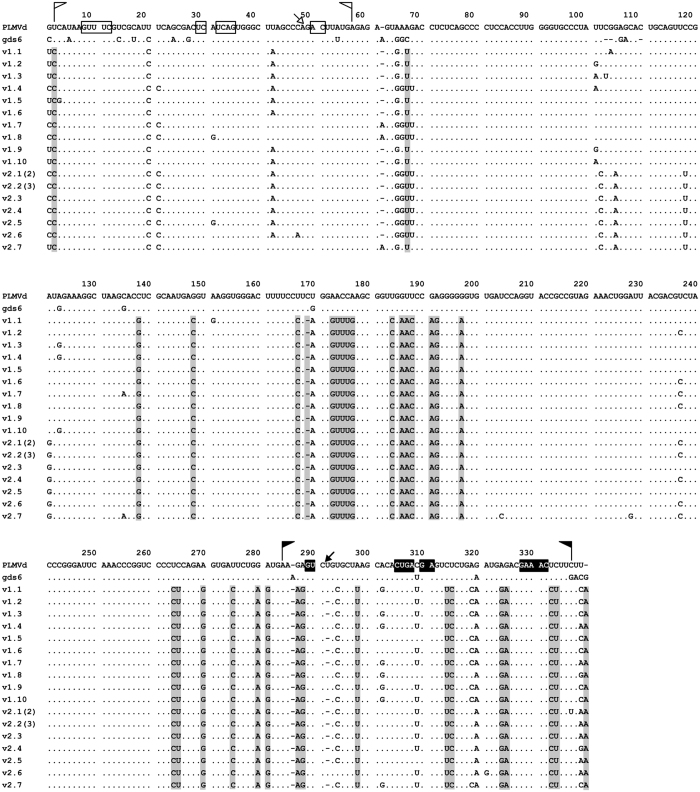
Figure 3. Alignment of the 17 different variants obtained from two PLMVd isolates of class II (V1 and V2) by RT-PCR using the pair of adjacent primers of opposite polarity RF1251 and RF1252 (Supplementary Table S1).
The reference and gds6 variants (of class I), included for comparative purposes, appear at the top. Dashes denote gaps and dots nucleotide identity with respect to the reference sequence. Regions forming the plus and minus hammerhead structures are flanked by flags, the nucleotide residues conserved in most natural hammerhead structures of viroid and viroid-like satellite RNAs are within boxes, and the self-cleavage sites are shown by arrows; black and white backgrounds refer to plus and minus polarities, respectively. Informative positions of the 17 variants of class II, with respect to the reference and gds6 variants of class I, are with grey background. When the same variant was recovered more than once it is indicated between parentheses. Other details as in the legend to Fig. 2.