Fig. 3.
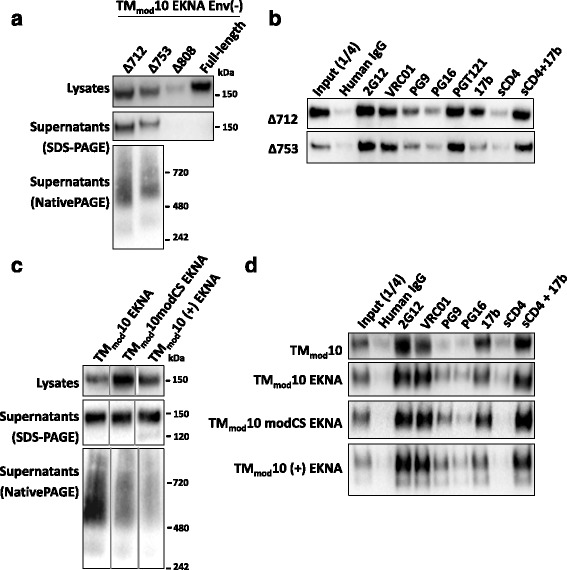
Effects of cytoplasmic tail and cleavage site modifications on TMmod10 Env. a The TMmod10 EKNA variant has the E168K + N188A changes that allow the HIV-1JR-FL Env to be recognized by the PG9 and PG16 antibodies [96–99]. The full-length TMmod10 EKNA Env or the TMmod10 EKNA variants with a deleted (Δ712) or truncated (Δ753 and Δ808) cytoplasmic tail were expressed in 293T cells. The cell lysates and supernatants were analyzed by SDS-PAGE, and the cell supernatants by Blue Native PAGE. The gels shown in this figure were Western blotted with a polyclonal goat anti-gp120 antibody. Addition of the cytoplasmic tail did not prevent TMmod10 EKNA expression in cells, but only the TMmod10Δ753 EKNA glycoprotein with the shortest tail was secreted. The secreted TMmod10Δ753 Env was mainly dimeric based on the Blue Native gel. b The TMmodΔ712 and TMmodΔ753 EKNA Envs precipitated from the supernatants of expressing cells by the indicated antibodies are shown. The antigenic profiles of these two Envs are similar. c TMmod10 EKNA variants with modifications of the cleavage site, including a wild-type cleavage site (+) or a flexible linker ((GGS)4) replacing the cleavage site (modCS), were analyzed as in a. Note that the TMmod10 (+) EKNA Env is partially cleaved, as indicated by the presence of a gp120 band on SDS-PAGE. d Precipitation of the TMmod10 and TMmod10 EKNA variants by the indicated antibodies is shown. Data are representative of those obtained in at least two independent experiments
