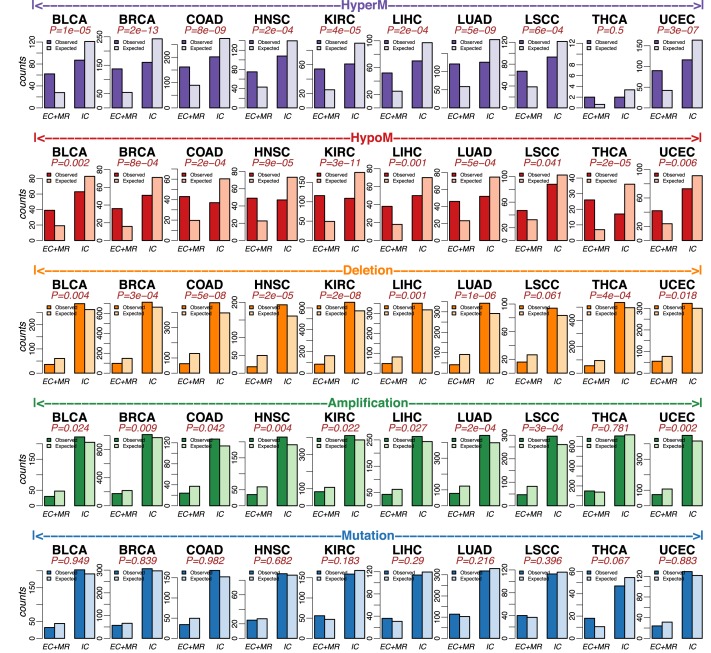
Figure 2.
Functional epigenetic alterations preferentially target genes in the extra-cellular/transmembrane domains, with SCNA counterparts preferentially mapping to the intra-cellular domain. Top Row: Barplots comparing the observed and expected numbers of genes for two different signaling domains (extracellular+transmembrane receptor: EC+MR, and intra-cellular: IC) for HyperM group (hypermethylated and underexpressed genes) across 10 TCGA cancer types. P-values are from a one-tailed Fisher's exact test. Alternative hypothesis is that the odds ratio of finding more genes mapping to the EC+MR domain is >1. Row-2: As before but for the HypoM group (hypomethylated and overexpressed genes). P-values are from a one-tailed Fisher's exact test. Alternative hypothesis is that the odds ratio of finding more genes mapping to the EC+MR domain is >1. Middle Row: As before but for the Deletion group (SCN deletion and underexpressed genes). P-values are from a one-tailed Fisher's exact test. Alternative hypothesis is that the odds ratio of finding more genes mapping to the EC+MR domain is <1. Second last row: As before but for Amplification group (SCN gain and overexpressed genes). P-values are from a one-tailed Fisher's exact test. Alternative hypothesis is that the odds ratio of finding more genes mapping to the EC+MR domain is <1. Last row: As before, but for Mutation group (mutated genes). P-values are from a one-tailed Fisher's exact test. Alternative hypothesis is that the odds ratio of finding more genes mapping to the EC+MR domain is >1.