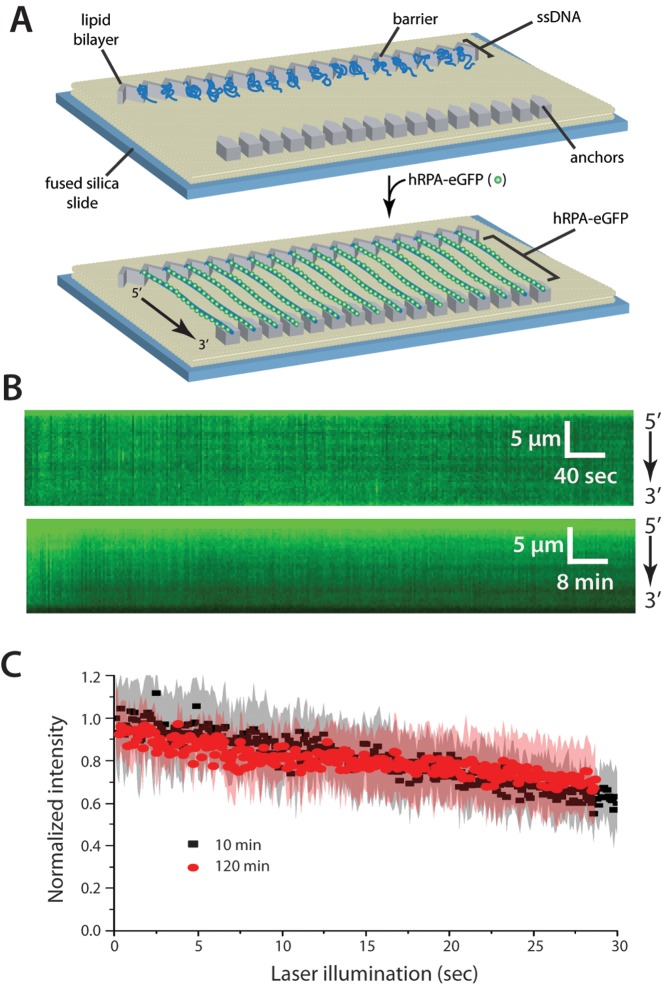
Figure 1.
Human RPA can bind tightly to ssDNA. (A) Schematic of the double-tethered hRPA-ssDNA curtain, showing the nanofabricated patterns on the surface of a fused silica microscope slide. The ssDNA molecules are anchored to the lipid bilayer in defined a 5′→3′ orientation through a biotin-streptavidin-biotin linkage and aligned at the zig-zag shaped chromium (Cr) barriers. The ssDNA is labeled and extended by injection of hRPA-eGFP and downstream ends are anchored through non-specific adsorption of the RPA-ssDNA to the exposed Cr pedestals. (B) Kymographs showing single ssDNA molecules bound by hRPA-eGFP in the absence of free RPA. Images (100-msec exposure) where collected at 2-s intervals for 10 min (upper panel) or at 24-s intervals for 2 h (lower panel), as described (26). (C) Loss of RPA-eGFP signal over time is due to photobleaching. Collecting images with longer shutter time (24 s) led to the same rate of signal decrease when corrected for total illumination time. Each curve represents normalized averages over time calculated from at least 14 individual ssDNA molecules, and the shaded regions represent standard deviation.