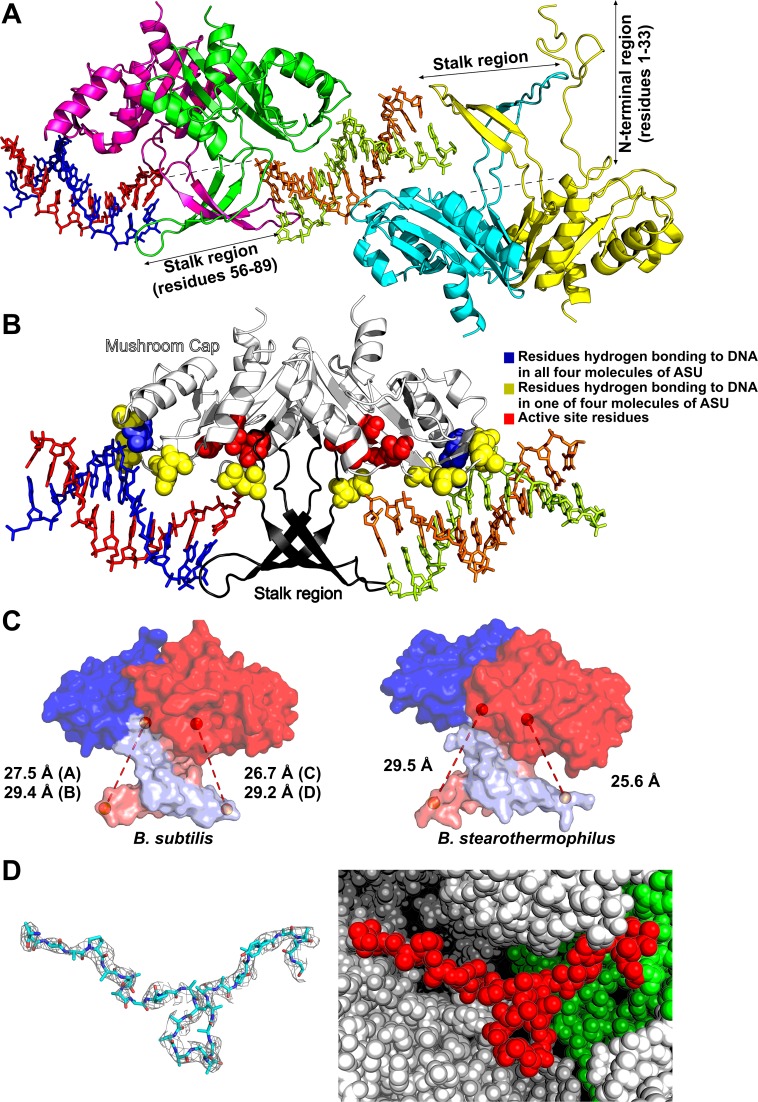
Figure 1.
Crystal structure of inactive mutant of RecU (RecUD88N)–DNA complex. (A) Crystallographic asymmetric unit (ASU) in crystal structure of RecU–DNA complex consists of four RecU monomers forming two dimers bound to two DNA duplexes. Cartoon representation of protein Chains A, B, C and D are shown in green, cyan, pink and yellow respectively. Chain E, F, G and H of DNA are shown in orange, limon, blue and red respectively. (B) Interactions of RecU with DNA duplexes: Cap and stalk regions are shown in white and black, respectively. Hydrogen bonding and active site residues are shown as van der walls spheres, while the DNA is shown as stick. Residues which form hydrogen bonds in all four monomers of RecU in ASU are shown in blue. Residues that form hydrogen bonds in only some of monomers are shown in yellow. Active site residues are shown in Red. (C) Distance between Cα atoms of residues R71 of one monomer and D88 of its dimerising partner (binding pocket) for both AC and BD dimer are shown for the structure presented here and for the structure of B. stearothermophilus. (D) Electron density (2Fo – Fc map) is shown at sigma level of 0.7 for the 33 residue long NTR region of chain D. the sphere representation is shown for the NTR coloured red. The ASU is coloured green while symmetry mates forming the crystal contacts are coloured white.