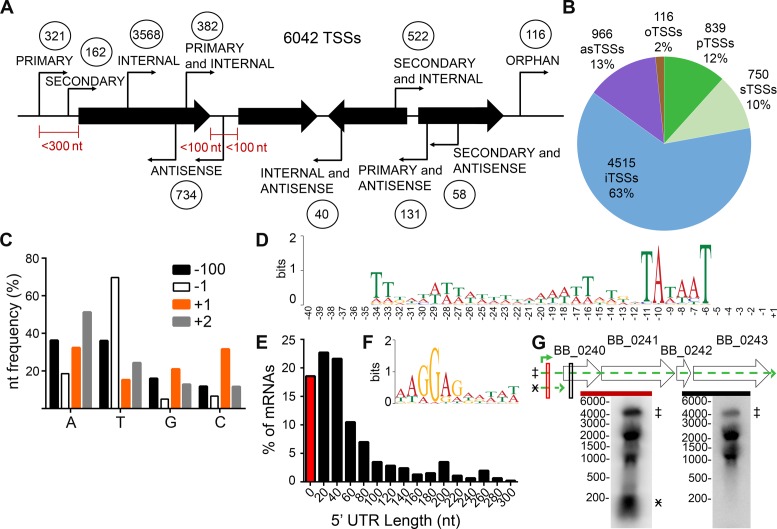
Figure 1.
Genome-wide identification and characterization of B. burgdorferi transcription start sites (TSSs). RNA was isolated from log phase B. burgdorferi clone B31 A3, and treated with and without TAP (tobacco acid phosphatase). (A) Schematic classification of the 6042 TSSs relative to the genome organization. Circled numbers indicate TSSs for each category. The maximum nucleotide distances from the 5′ and/or 3′ of annotated sequences, shown as black arrows, for a particular category of TSS are indicated in red. (B) Graphical representation of the TSS classifications. as, antisense; o, orphan; p, primary; s, secondary; i, internal. (C) Nucleotide frequency at the −100, −1, +1 (TSS) and −1 nucleotide positions. A, adenine, T, thymine, G, guanine, C, cytosine. (D) Consensus motif for promoter regions upstream of primary TSSs. The nucleotide sequences from −40 to +1 of the 321 identified primary TSSs were analyzed by MEME 4.11.2. (E) Distribution of 5′ untranslated regions (UTR) lengths among uniquely classified primary and secondary TSSs. The data are shown as the percent of mRNA sequences with a 5′ UTR of each bin length. 0–10 nucleotides (red bar), bin size of 20 nucleotides, where the number shown is the middle length in each bin (black bars). (F) Consensus ribosome binding motif for uniquely classified, primary 5′ UTRs. MEME 4.11.2 was used to analyze UTR sequences ranging from 10–293 nts in length for a conserved motif, which was found on average to begin 8 nts upstream of the annotated start codon. (G) Northern blot analyses validate a long 5′ UTR. Total RNA was extracted from mid-log phase spirochetes and separated by denaturing formaldehyde–agarose gel, blotted to nylon membranes and probed with 32P-labeled complementary probes, indicated by red and black boxes. Genomic context of ORFs BB_0240-BB_0243 (wide black arrows), the primary TSS (green bent arrow), putative transcripts and their position on the Northern blot (broken line, green arrows and marked with designated symbol) are indicated. Marker sizes in nucleotides are indicated to the left of each blot.