Figure 1.
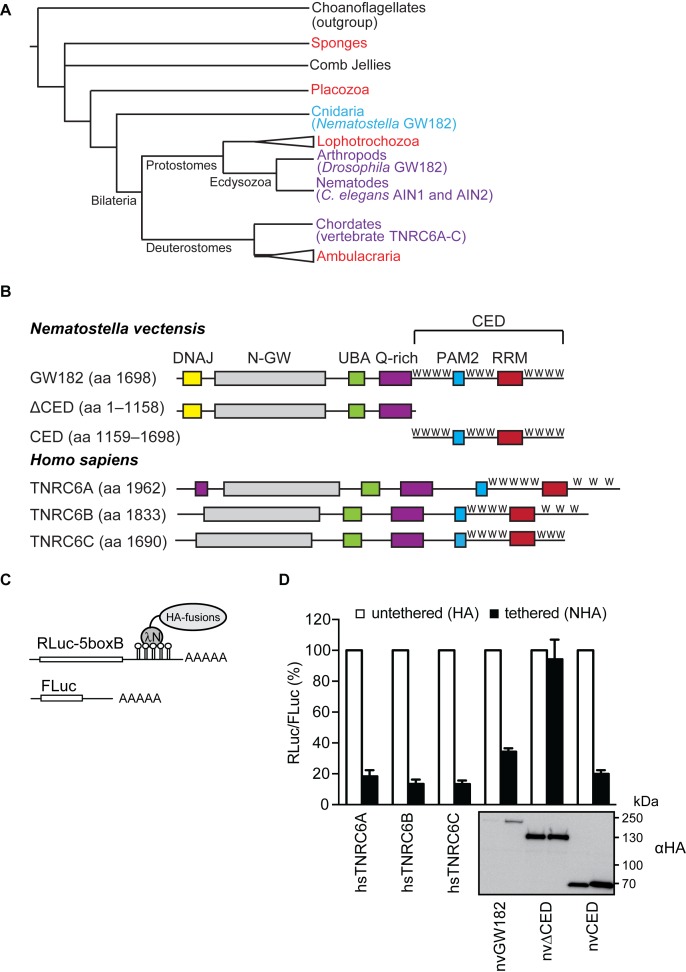
nvGW182 represses tethered mRNA via its C-terminal effector domain (CED). (A) Schematic phylogenetic tree of Metazoa at the phylum level [based on (53,54)]. Phyla where GW182 proteins were functionally and biochemically studied appear in purple and the studied protein appear within brackets. Phyla where GW182 homologs are found in the genome but have not been functionally studied yet appear in red. Cnidaria appear in light blue. Phylogenetic groups of multiple phyla are indicated by a triangle. The illustrated polytomy of sponges and comb jellies is due to the current uncertainty regarding their relative phylogenetic positions. (B) Schematic representation of Nematostella vectensis GW182 (nvGW182), its human homologs TNRC6A, B and C, and their deletion mutants. DNAJ domain (yellow); N-GW: GW-repeat–rich region (grey); UBA: ubiquitin associated domain (green); PAM2: PABP associated motif 2 (blue); RRM: RNA-recognition motif (red). The C-terminal effector domain (CED) is formed RRM, PAM2 and the unstructured flanking regions with tryptophan-containing motifs, or W-motifs (W). The numbers correspond to the amino acid positions. (C) Schematic representation of reporter constructs used in tethering experiments: RLuc-boxB encodes for Renilla luciferase (RLuc) coding sequence fused to a 3′UTR with five boxB sites that specifically bind to λN peptide; FLuc encodes for Firefly luciferase (FLuc) coding sequence without boxB sites and serves as a control (55). (D) Repression of RLuc-boxB mRNA by NHA-nvGW182 and its deletion mutants. Human HEK293 cells were co-transfected with plasmids encoding RLuc-boxB, FLuc and full-length NHA-nvGW182 or indicated NHA-nvGW182 deletion mutants. As positive controls, plasmids encoding human NHA-TNRC6A, NHA-TNRC6B and NHA-TNRC6C were co-transfected. As negative controls, plasmids encoding untethered HA-fusions were used. RLuc activity was normalized to that of FLuc and presented as a percentage of RLuc produced in the presence of the corresponding untethered HA-fusions (open bars). Values represent means ±SD from 5 to 6 experiments. Expression levels of HA-fusion proteins were estimated by Western blotting with antibodies directed against HA-peptide.