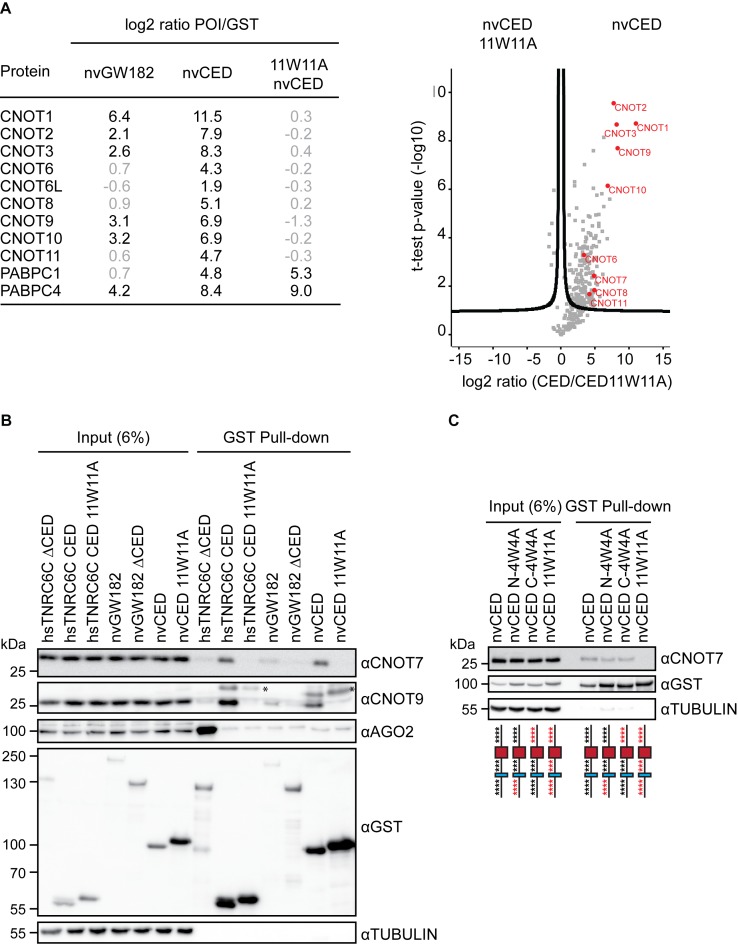
Figure 3.
nvGW182 recruits subunits of the CCR4-NOT complex via its W-motifs. (A) Quantitative mass spectrometric analysis of proteins associated with nvGW182. The GST-nvGW182 full-length constructs, the nvCED domain only, as wild type and 11W11A mutant variant, were expressed in HEK293 cells and affinity purified using GST. GST alone was used as a negative control. On the right, relevant interactors for each variant are listed along with their corresponding enrichment factor (the log2 ratio of label-free quantification values (LFQ intensity) over corresponding LFQs detected in pull-down of GST tag alone). On the left, volcano plot showing proteins with differential binding to nvCED and nvCED11W11A variant. The logarithmic ratios of protein intensities were plotted against negative logarithmic P-values of a two samples t-test (two sided). A hyperbolic curve separates significantly enriched proteins from common binders. Selected interactors recruited via W-motifs of nvCED are labeled in red (FDR ≤ 0.05, n = 2). (B) Validation of CCR4-NOT-nvCED interactions identified in (A). GST-fusions of indicated proteins were expressed in HEK293 cells, and inputs (6%) and GST-pulldowns were analyzed by Western blotting using the indicated antibodies. Human TNRC6CΔCED was used as positive control for AGO2 binding. TNRC6C CED and TNRC6C 7W7A CED were used as positive and negative controls for CNOT7 and CNOT9 binding. Asterisks (*) indicate unspecific bands. (C) W-motifs function additively to recruit the CCR4-NOT complex. The experiment was performed as in (B), except that nvCED with four N-terminal (N-4W4A) or C-terminal (C-4W4A) W-motifs mutated were used for pull-downs. nvCED and nvCED 11W11A were used as positive and negative controls for CNOT7 binding, respectively. Schematic representations of nvCED domains are shown below the corresponding lane. Asterisks indicate W-motifs; W residues are shown in black and W→A mutations in red.