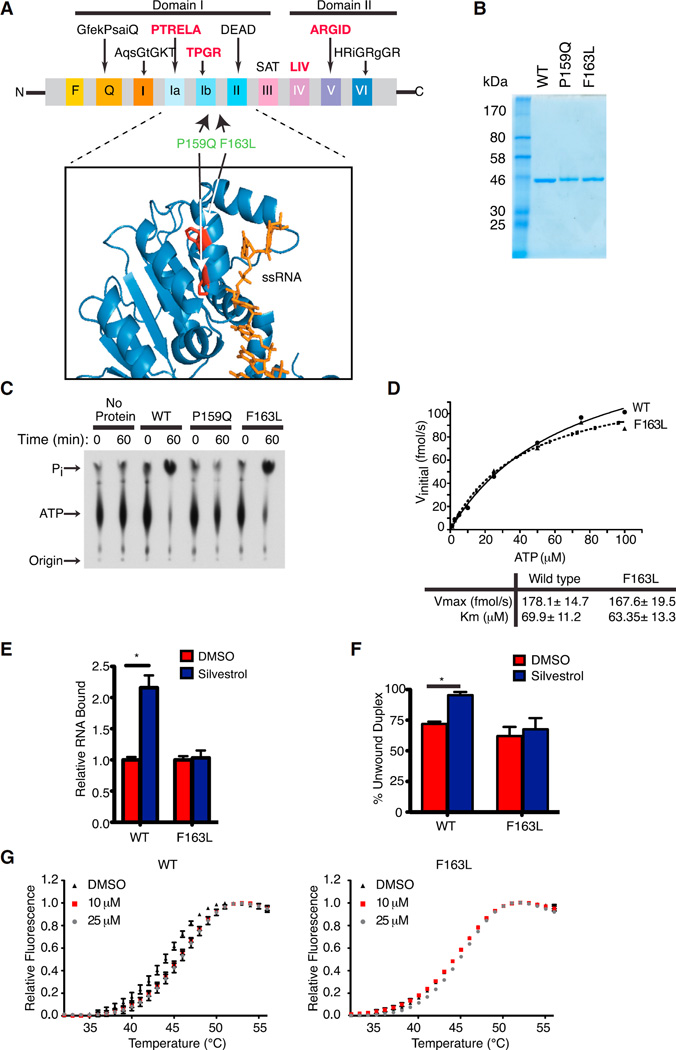
Figure 1. Generation and Characterization of a Rocaglate-Resistant eIF4A1 Allele.
(A) Schematic illustrating conserved motifs of the DEAD-box helicase family. Sequences of the conserved motifs are denoted with motifs involved in RNA binding highlighted in bold red. The structure of eIF4A indicates it to be dumbbell in shape with two domains (I and II) linked via a flexible linker sequence. The inset shows a ribbon diagram of eIF4A1 (PDB 2ZU6) aligned to eIF4A3 (not shown; PDB 2HYI). The residues targeted for mutagenesis are highlighted and the single-stranded RNA substrate (positioned relative to the eIF4A3 crystal structure) is shown in orange.
(B) Coomassie stain of purified recombinant eIF4A1 proteins.
(C) Assessment of ATP hydrolysis by recombinant proteins via thin layer chromatography.
(D) Kinetics of ATP hydrolysis by eIF4A1 and eIF4A1(F163L). ATPase assays were performed with 1 µg protein and varying ATP concentrations. Graph represents the Michaelis-Menten fit from two independent experiments.
(E) RNA binding activity of eIF4A1 and eIF4A1(F163L) using [32P]-labeled RNA generated from pSP/CAT (see Supplemental Experimental Procedures). Assays were performed in the presence of 0.5% DMSO or 1 µM silvestrol and the retained eIF4A:RNA complexes are set relative to DMSO controls. n = 3 biological replicates performed in triplicate ±SEM; *p < 0.001.
(F) Quantitation of eIF4A1 and eIF4A1(F163L) helicase activity performed with 0.5 µg recombinant eIF4A1 and an 11-nt radiolabeled RNA duplex in the presence of DMSO or 50 µM silvestrol. n = 3 biological replicates ±SEM; *p < 0.05.
(G) DSF analysis of eIF4A1 or eIF4A1(F163L) in the presence of DMSO or (−)-SDS-1-021.