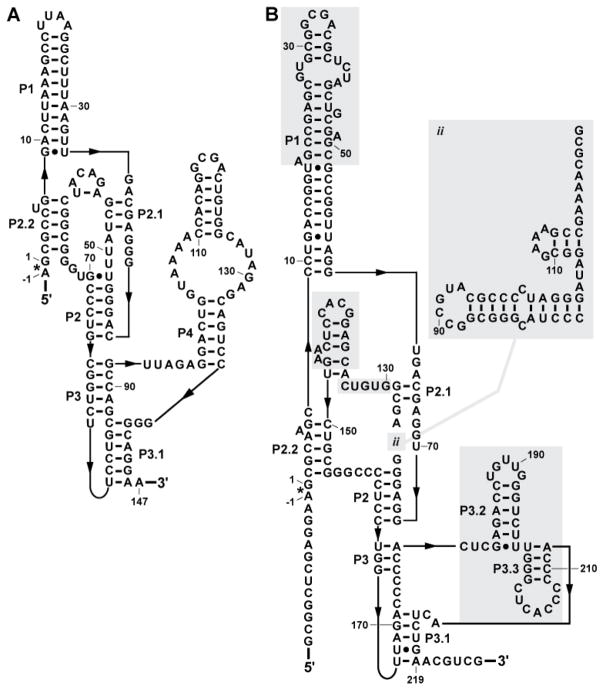
FIGURE 2.
Comparison of a canonical glmS ribozyme with an extreme variant. (A) The ribozyme from Thermoanaerobacter tengcongensis corresponds well to the consensus glmS ribozyme architecture typical of the vast majority of representatives. The asterisk identifies the site of self-cleavage and arrows denote zero-length connections. (B) A variant glmS ribozyme present in Truepera radiovictrix. Shaded regions identify sequences and structures that are substantially different from the typical glmS ribozyme consensus architecture.