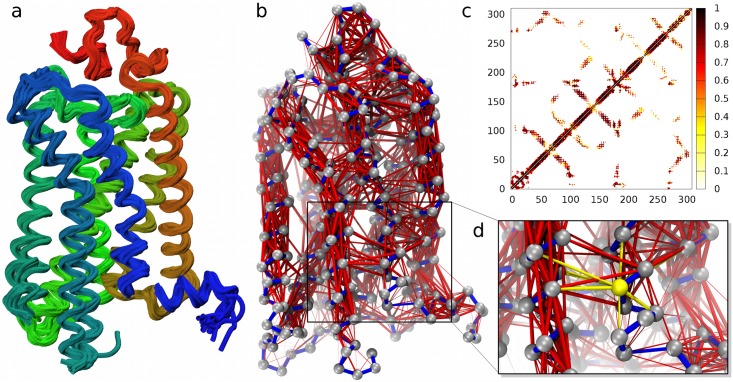
Fig 1. Scanning GPCRs’ mechanical network for key sites.
The structural ensemble of a G protein-coupled receptor, see panel (a) for rhodopsin, is used to compute the distance variations for all pairs of amino acids. (b) The pairings in the local mechanical network (Cα distance <12Å) are highlighted with red bonds with thickness proportional to the observed rigidity; only the strongest links are shown here, while the full network is shown in Fig A in S1 Supporting Information. The network is represented as a color-coded contact map in panel (c). Key residues for the overall mechanical integrity of the network are identified by measuring how the network connectedness varies when one removes all the links of a node corresponding to non-covalent bonds (highlighted in yellow in panel d).