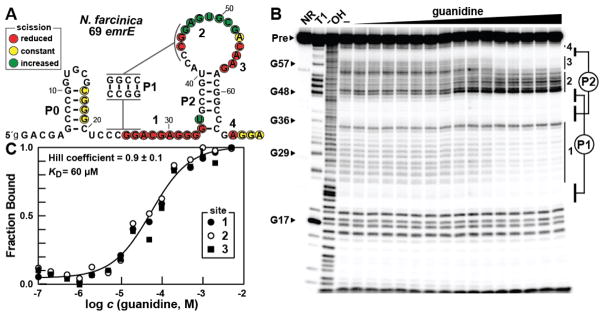
Figure 2.
Guanidine is bound by a guanidine-III RNA in vitro. (A) The sequence and observed secondary structure of the 69 emrE RNA from N. farcinica. Lowercase 5′ nucleotides indicate G residues added to enable transcription by T7 RNA Polymerase. Red, yellow, and green circles indicate nucleotides that undergo decreased, constant, or increased scission, respectively, upon addition of the ligand as determined by their relative band intensities in B. Regions of guanidine-mediated structural modulation numbered to correspond to those in B and C. (B) Polyacrylamide gel electrophoresis (PAGE) analysis of in-line probing reactions of 69 emrE RNA in the absence (−) and presence of guanidine ranging from 100 nM to 5 mM. NR, T1, and −OH indicate no reaction, RNA partially digested with T1 ribonuclease (cleavage after each G), and RNA partially digested under alkaline conditions (cleavage after every nucleotide). Precursor RNA (Pre) as well as certain G residues within the RNA sequence are indicated and regions of modulation are numbered and indicated with vertical bars. Base pairing of the pseudoknotted P1 and P2 stems is depicted. (C) Plot of the fraction of RNA bound to ligand versus the logarithm of the guanidine concentration as determined by quantification of B (see reference 12 for details). The KD and Hill coefficient were determined by the sigmoidal fit and the reported error represents the standard deviation reported in the fitting parameters.