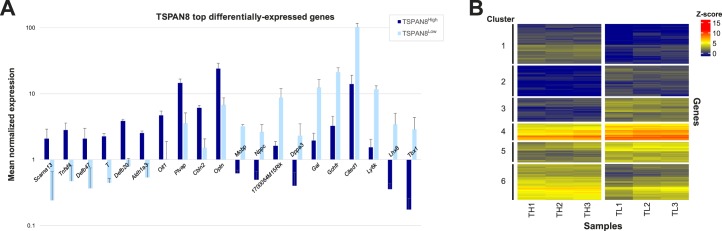
FIG. 5.
Differential gene expression between the TSPAN8High and TSPAN8Low subpopulations of P6 ID4-EGFP+ spermatogonia. Significant differences were observed in levels of genes with an average expression value of >2 between the ID4-EGFP+/TSPAN8High and ID4-EGFP+/TSPAN8Low spermatogonia populations. Genes with statistically significant differences in levels between populations (P < 0.0001) and at least 2-fold difference in levels between ID4-EGFP+ spermatogonial populations were considered to be differentially expressed. A) The top differentially expressed genes (fold-change) are shown for comparisons between TSPAN8High and TSPAN8Low. Graphs portray the mean ± SD mRNA levels (normalized expression counts) of the noted genes for each ID4-EGFP+ subpopulation. All bar pairs are significantly different (P < 0.0001) between the TSPAN8High and TSPAN8Low subpopulations. B) Hierarchical clustering using only the differentially expressed genes was used to confirm separation between individual sample replicates and group differentially expressed genes. Heatmaps of global Z-score for the differentially expressed genes are shown for comparisons between the three replicates each of the ID4-EGFP+/TSPAN8High (TH1, TH2, TH3) and ID4-EGFP+/TSPAN8Low (TL1, TL2, TL3) subpopulations. GO analyses of these differentially expressed genes are found in Table 1 and Supplemental Table S8. Horizontal clustering (samples) is based on Euclidean distance (dendrogram not shown) and vertical clustering (genes) is based on Spearman correlation coefficient. K means clustering (six clusters) was then used to group genes with like expression patterns. Cluster labels (1–6) denote gene groups that can be found in Supplemental Table S7. GO analyses of clusters are found in Supplemental Table S10. Note: a parallel analysis with 1.5-fold differences in mRNA abundance is shown in Supplemental Figure S5 and Supplemental Table S9.