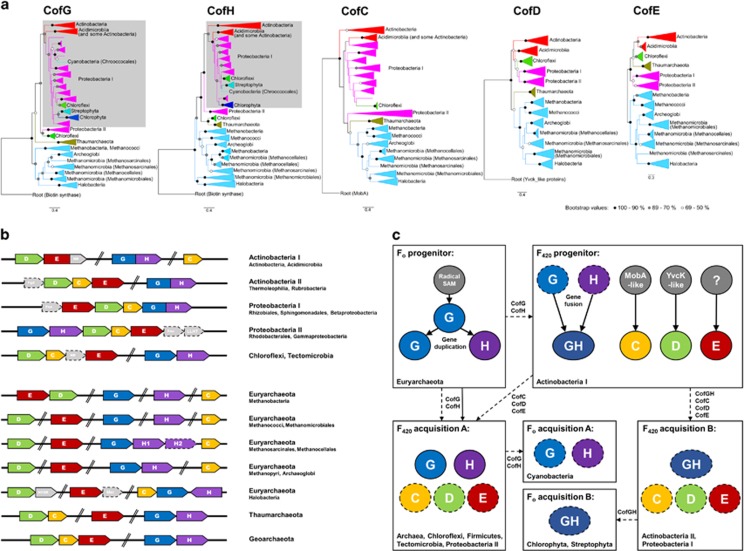
Figure 4.
Evolution of the determinants of F420 biosynthesis. (a) Phylogenetic trees of the F420 biosynthesis proteins (CofC, CofD, CofE, CofG and CofH). Trees were rooted with a related protein family characterized by the same protein fold, except for CofE (novel protein fold), presented as an unrooted tree. Clades are labeled according to phyla, except for Actinobacteria and Euryarchaeota that are labeled by class. The Firmicutes, Tectomicrobia, Thermoleophilia and Rubrobacteria lineages have been omitted as their inclusion compromises phylogenetic inferences because of long-branch attractions. Gray-shaded regions represent the CofGH fusion proteins. CofG and CofH trees incorporating cyanobacterial sequences are presented in Supplementary Figure S4 as these sequences caused low bootstrap values at key nodes. (b) Generalized schematic of the genetic organization of the genes encoding the five enzymes specifically required for F420 biosynthesis (CofC, CofD, CofE, CofG and CofH) from five bacterial phyla (Actinobacteria, Proteobacteria, Chloroflexi, Tectomicrobia and Firmicutes) and three archaeal phyla (Euryarchaeota, Thaumarchaeota and Geoarchaeota). Fgd, F420-reducing glucose 6-phosphate dehydrogenase; Fno, F420-reducing NADPH dehydrogenase; HFDR, predicted F420H2-dependent reductase; LLHT, predicted F420H2-dependent luciferase-like oxidoreductase; Mer, methylenetetrahydromethanopterin reductase; NR, hypothetical nitroreductase. (c) Schematic representation of the proposed evolutionary origin of F420 biosynthesis genes and their acquisition by different phyla. Solid lines/circles indicate vertical acquisition, whereas dashed lines/circles indicate horizontal acquisition. The capacity for F420 biosynthesis appears to have evolved on at least two separate occasions in both the Actinobacteria (Actinobacteria I=Actinobacteria, Acidimicrobiia; Actinobacteria II=Thermoleophilia, Rubrobacteria) and Proteobacteria (Proteobacteria I=Rhizobiales, Sphingomonadales, Betaproteobacteria; Proteobacteria II=Rhodobacterales, Gammaproteobacteria).