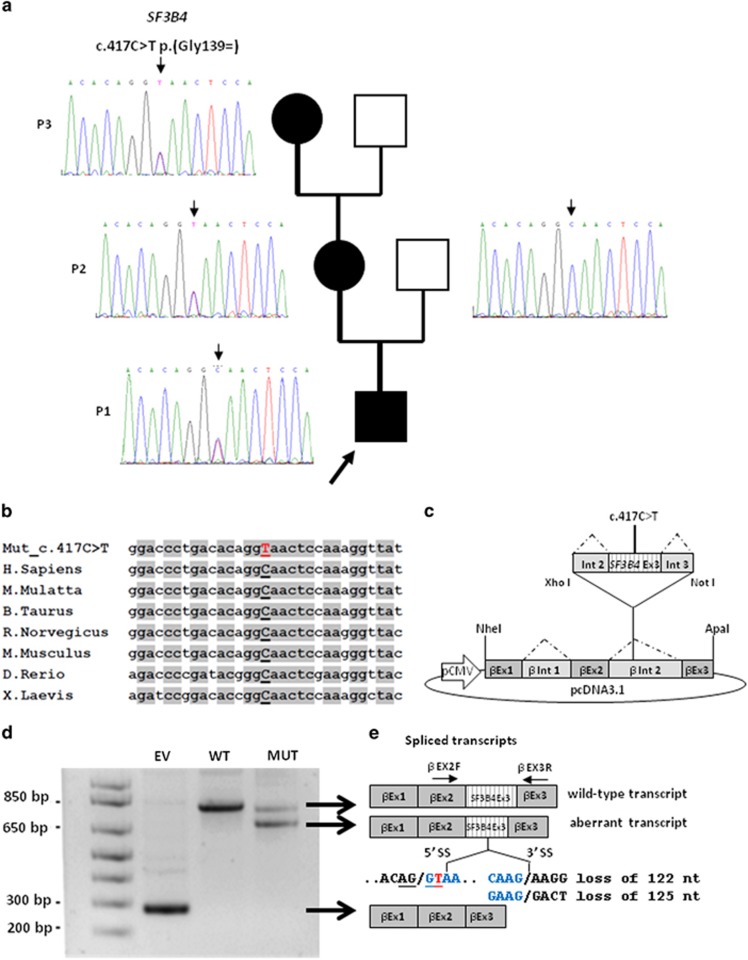
Figure 2.
Genetic findings in our family with Nager syndrome. (a) SF3B4 sequencing shows the heterozygous c.417C>T substitution in P1, P2 and P3, and the wild-type allele in the unaffected proband's father. (b) The c.417C>T variant affects a highly conserved nucleotide throughout evolution. (c) Schematic representation of the β-globin minigene construct used in this study. βEx1, βEx2, βEx3 refer to β-globin exon 1, 2 and 3. βInt1, βInt2 refer to β-globin intron 1 and 2. Wild-type and mutant exon 3 (SF3B4 Ex3) with at least 100 bp of the flanking introns (Int 2 and Int 3) were cloned into β-globin intron 2. (d) Total RNA extracted from cells transfected with the wild-type (WT), the mutant construct (MUT) and the empty vector (EV) was retro-transcribed and PCR-amplified using primers specific for β-globin exon 2 (βEX2F) and 3 (βEX3R) and separated by agarose gel electrophoresis. The mutant construct (MUT) showed an aberrant splicing compared with controls (WT) and yielded two bands. (e) Schematic representation of the structure of the transcripts obtained from the hybrid minigenes. The shorter transcript lacks an interstitial portion of exon 3 owing to the activation of a novel donor splice site (5′ SS) and the use of two different acceptor splice sites (3′ SS). The first and last nucleotides of the missing portion of exon 3 are indicated in blue and the novel SF3B4 pathogenic variant is highlighted in red. The upper band corresponds to the correctly spliced transcript.