Sir,
Clinical cytogenetics aims at the identification of chromosomal abnormalities by microscopic observation of metaphase chromosomes, known as karyotyping. Technical innovations such as arrays have replaced karyotyping as the first tier test for the identification of pathogenic genomic imbalances in referrals of intellectual disability and congenital anomalies.1, 2 It is likely that in the near future, whole-genome sequencing will become the first tier test for such patients, which has the advantage of combining in a single test the identification of pathogenic mutations and imbalances.3 These exciting developments have caused a growing need for technicians and laboratory geneticists with specialized training in molecular genetics, and a shift from the microscope to DNA sequencing and interpretation of DNA variants.
The hidden danger of this shift is that the competency of laboratories offering cytogenetic services may fade away. Cytogenetic analysis will still be required as a second line test where the mechanism leading to the chromosomal rearrangement or clonal abnormalities need to be ascertained for patient and/or family management. The 2016 online external quality assessment (EQA) of postnatal karyotyping by the Cytogenetic EQA Service (CEQAS, www.ceqas.org) now provides the first evidence that this danger is imminent and will lead to incorrect clinical diagnoses. The case in point concerned a male who was referred for karyotyping because of hypogonadism and infantile genitalia. He had a mosaic karyotype with about equal proportions of 46,X,idic(Y)(q11.?21) and 45,X metaphases in lymphocytes from peripheral blood. As can be seen in Figure 1, the Y chromosome was unusually short and looked symmetric. Fluorescence in situ hybridization showed that two copies of the SRY gene were present, located at the terminal ends of each arm of the idic(Y), and that two DYZ3 loci (centromeres) were present. There was no signal for DYZ1 (band Yq12) in 200 interphase nuclei investigated. Thus, there was no evidence for a normal Y chromosome in this male. Nevertheless, 48 of the 164 laboratories (29.3%) that participated in this EQA reported a 45,X/46,XY mosaic karyotype, and 3 laboratories (1.8%) reported a del(Y)(q11.?21) instead of idic(Y)(q11.?21). In total, 51 participants (31.1%) failed to identify the abnormality correctly, an unprecedented high number.
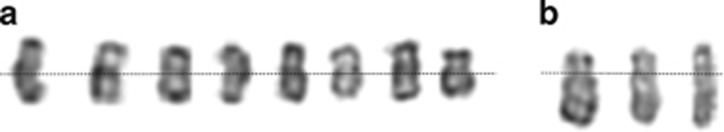
Figure 1.
The idic(Y)(q11.?21) chromosome from a male with hypogonadism that was mistaken by ~30% of participating laboratories for a normal Y chromosome. The karyotype was 46,X,idic(Y)(q11.?12)[17/]/45,X[13].ish idic(Y)(SRY++).nuc ish(DYZ3x2)[60/100],(DYZ1x0)[200]. (a) Representative idic(Y) chromosomes, each from a different metaphase. The dashed line represents the axis of symmetry. (b) For comparison, a structurally normal Y chromosome is shown. The dashed line is at the position of the centromere.
An interpretation of this case as 45,X/46,XY causes an incorrect clinical diagnosis to be made, since it leads to the wrongful assertions that the male fertility genes on the long arm of the Y chromosome are present and that spermatogenesis would be possible. The abnormal Y chromosome contains two copies of the short arm and the proximal part of the long arm of the Y chromosome and lacks the more distal long arm material. In this particular example both array and DNA sequencing-based methods are unable to correctly identify the genomic abnormality because a distinction between a mosaic idic(Y) and a non-mosaic del(Y) would be impossible. In addition, the presence of 45,X cells could only be detected by microscopic observation of multiple, individual metaphases. Such idic(Y) chromosomes arise de novo by crossover between sister chromatids in the long arm of the Y-chromosome. The crossover takes place within one of the eight long palindromic DNA sequences in the long arm, each sharing >99% sequence identity.4 Thus, current methods for whole-genome paired-end sequencing would probably miss the breakpoint because of the repetitive nature of the surrounding DNA sequences. The position of the exchange defines the axis of symmetry within the idic(Y), and also the extent of loss of genes on the long arm that are required for spermatogenesis. In this particular idic(Y)(q11.?12) chromosome the point of symmetry in the long arm is relatively close to the centromere, leading to infertility through loss of AZFb and AZFc, and perhaps also AZFa.5 This explains the hypogonadism in the patient and leads to the conclusion that spermatogenesis is absent. Therefore, it is inappropriate that several participating laboratories stated that this patient could have children through the application of artificial reproduction techniques.6, 7
The fact that one in three laboratories failed this test is alarming evidence that cytogenetic proficiency has started to fade away. The basic skill that was on test in this EQA was the identification of a structurally abnormal G-banded chromosome, not the detection of a tiny deletion at the lower range of microscopic resolution. The morphological evaluation of human metaphase chromosomes requires a specialized training period of several years during which an adequate number of cases must be analyzed (see Appendix for examples). This assures that aspiring laboratory specialists are familiar with the human karyotype and are able to correctly identify any rare, exceptional chromosomal abnormality. In addition, they must gather sufficient experience to be competent to report such cases according to international guidelines.8 The training required for registration as a clinical laboratory geneticist varies between countries. For example, the training program of the Canadian College of Medical Geneticists (www.ccmg-ccgm.org) requires that at least 100 cases are analyzed in the laboratory and over 200 cases are evaluated and reported during a 2-year period. Also in the United States of America a specialized 2–3 year training program exists.9 In the United Kingdom there is a combined 3-year training program that includes basic cytogenetics, molecular genetics and some aspects of pathology. In The Netherlands, a 4-year training program, including the reporting of a minimum number of 1000 cases, was replaced in 2013 by a program that combines cytogenetics, molecular genetics, biochemical genetics and oncogenetics as one profession (www.vkgl.nl). This novel program has been approved by the European Society of Human Genetics (www.eshg.org) as a template for other countries of the European Union,9 but allows for only 3 months of training in the cytogenetic laboratory without specifying a minimum number of cases that must be analyzed and reported (www.vkgl.nl). The result of the 2016 postnatal bloods EQA rings an alarm bell for cytogenetic competency. Ensuring the competencies of laboratory personnel is an important part of quality assurance. Laboratory directors and board members of societies for human genetics now face the responsibility to ensure that sufficient numbers of candidates enter the training programs, and that the competencies of staff members involved in karyotyping are ensured. Our finding also provides evidence for the importance of participating in schemes for EQA.
Acknowledgments
We are indebted to Bettina Quellhorst-Pawley for expert technical and organizational skills, and we thank Wigard Kloosterman for discussion about the limitations of paired-end sequencing methods.
Appendix
Internet resources for training programs in clinical cytogenetics
Canada
Canadian College of Medical Geneticists. https://www.ccmg-ccgm.org/images/Training/Cytogenetics/Cytogenetic_Training_Guidelines_2014.pdf
European Union
European Society of Human Genetics/European Board of Medical Genetics. https://www.eshg.org/fileadmin/eshg/EBMG/CLG/Core-Curriculum_2015.pdf
The Netherlands (since 2013)
Vereniging voor Klinisch Genetische Laboratorium Diagnostiek. http://www.vkgl.nl/docs/Opleidingseisen_laboratoriumspecialist_Klinische_Genetica.pdf
The Netherlands (until 2013)
Vereniging voor Klinisch Genetische Laboratorium Diagnostiek. http://www.vkgl.nl/docs/Samenvatting_specifieke_opleidingseisen_oude_stijl.pdf
United Kingdom
Association for Clinical Cytogenetics. http://www.cytogenetics.org.uk/training/clinical_scientist_training_programme.html
United States of America
American Board of Medical Genetics and Genomics. http://www.abmgg.org/pages/training_options.shtml
Footnotes
The authors declare no conflict of interest.
References
- Hochstenbach R, van Binsbergen E, Engelen J et al: Array analysis and karyotyping: workflow consequences based on a retrospective study of 36,325 patients with idiopathic developmental delay in the Netherlands. Eur J Med Genet 2009; 52: 161–169. [DOI] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S et al: Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010; 86: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehir-Kwa JY, Pfundt R, Veltman JA: Exome sequencing and whole genome sequencing for the detection of copy number variation. Expert Rev Mol Diagn 2015; 15: 1023–1032. [DOI] [PubMed] [Google Scholar]
- Lange J, Skaletsky H, van Daalen SK et al: Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell 2009; 138: 855–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausz C, Chianese C, Giachini C, Guarducci E, Laface I, Forti G: The Y chromosome-linked copy number variations and male fertility. J Endocrinol Invest 2011; 34: 376–382. [DOI] [PubMed] [Google Scholar]
- Brandell RA, Mielnik A, Liuotta D et al: AZFb deletions predict the absence of spermatozoa with testicular sperm extraction: preliminary report of a prognostic genetic test. Hum Reprod 1998; 13: 2812–2815. [DOI] [PubMed] [Google Scholar]
- Krausz C, Quintana-Murci L, McElreavey K: What is the clinical prognostic value of Y chromosome microdeletion analysis? Hum Reprod 2000; 15: 1431–1434. [DOI] [PubMed] [Google Scholar]
- Claustres M, Kožich V, Dequeker E et al: Recommendations for reporting results of diagnostic genetic testing (biochemical, cytogenetic and molecular genetic). Eur J Hum Genet 2010; 22: 160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yu J, Ming Q, Bao L, Wu B-L, Li P: On the globalization and standardization of medical genetics and genomics as clinical and laboratory specialities. N A J Med Sci 2014; 7: 194–198. [Google Scholar]