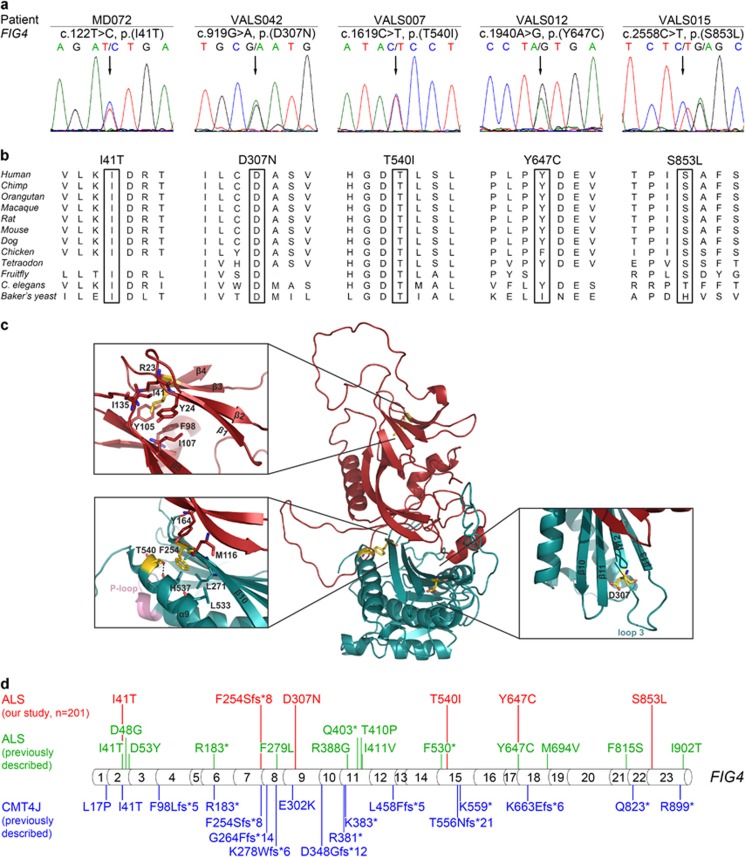
Figure 2.
Rare heterozygous deleterious FIG4 missense variants in five sALS patients detected by whole-exome and targeted sequencing. (a) Electropherograms demonstrating rare heterozygous FIG4 missense variants in five sALS patients (affected nucleotides are designated by an arrow). All variants are predicted to be deleterious by at least two prediction tools. Parents were not available for segregation analysis. (b) Missense variants detected in FIG4 affect highly or very highly conserved amino acids (data taken from Alamut Visual 2.6.1., Interactive Biosoftware, Rouen, France). (c) Mapping of the amino-acid residues affected by the ALS-associated variants detected here on a structural model of human phosphoinositide 5-phosphatase FIG4. One of the affected residues (p.I41) is located in the N-terminal SAC domain (red), and three affected residues (p.F254; p.D307; p.T540) are situated in the catalytic domain (cyan). The identified frameshift variant, FIG4:c.759delG, p.(F254Sfs*8), is predicted to result in a truncated protein lacking the catalytic domain (cyan) including the P-loop (active center). The insets show close-up views of the corresponding regions. The structural model was generated using MODELLER32 based on the crystal structure of SAC1 (PDB: 3LWT).20 There are no structural data covering the regions encompassing the residues p.Y647 and p.S853, which are, therefore, missing in the model. (d) Summary of FIG4 variants detected in ALS patients in this or previous studies or previously in CMT4J patients. Variants found in ALS patients from our cohort are indicated in red; variants previously described in ALS patients6, 19, 33 are indicated in green; variants previously described in CMT4J patients9, 16 are indicated in blue. Protein sequence variants are given according to the Human Genome Variation Society recommendation v2.0; the exon structure of the human FIG4 gene (NG_007977.1, NM_014845.5) was based on Alamut Visual 2.6.1.