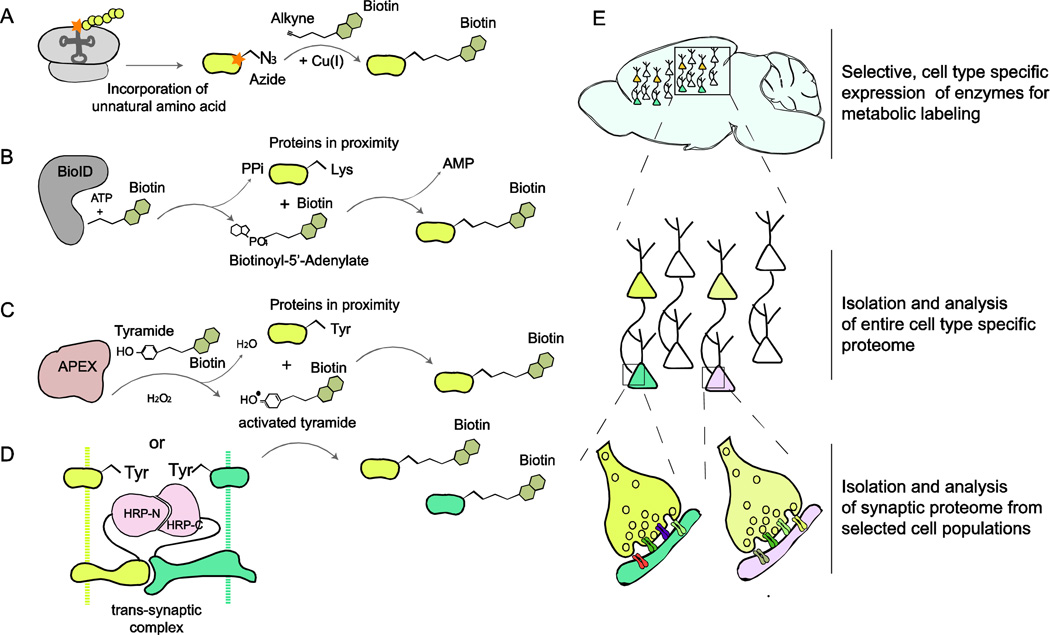
Figure 2. Overview of chemical labeling techniques and their application for targeted proteome analysis.
(A) Bioorthogonal noncanonical amino acid tagging (BONCAT): the approach is based on the in vivo incorporation of non-canonical, azide containing amino acids such as L-azidohomoalanine (AHA) into newly synthesized proteins. These proteins can be subsequently labeled with biotin by ‘click-chemistry’ and isolated for further analysis. (B) In vivo proximity protein biotinylation by promiscuous biotin-ligase (BioID): biotinylation occurs through Biotinoyl-5’-Adenylate which is released by a mutated variant of E.coli biotin ligase BirA. Biotinoyl-5’-Adenylate is a highly reactive compound that quickly reacts with lysines of proximal proteins. (C) Biotin-labeling by ascorbate-peroxidase (APEX) and (D) by split-horseradish-peroxidase: both techniques utilize the ability of peroxidase enzymes to generate highly reactive species from tyramide derived compounds such as biotin-phenol. These react quickly with aromatic groups (usually tyrosine and tryptophan, but also histidine and cytosine) of proteins in close proximity. (E) Application of metabolic labeling for analysis of cell-type/compartment-specific proteomes: Selective expression of biotin-labeling enzymes in cells of interest (e.g. utilizing the Cre-Lox-system) allows isolation of proteins expressed in identified cells. Additionally, targeting these enzymes to synaptic compartments provides an opportunity for the labeling and subsequent determination of synapse-type specific protein composition in desired cell populations.