Fig. 1.
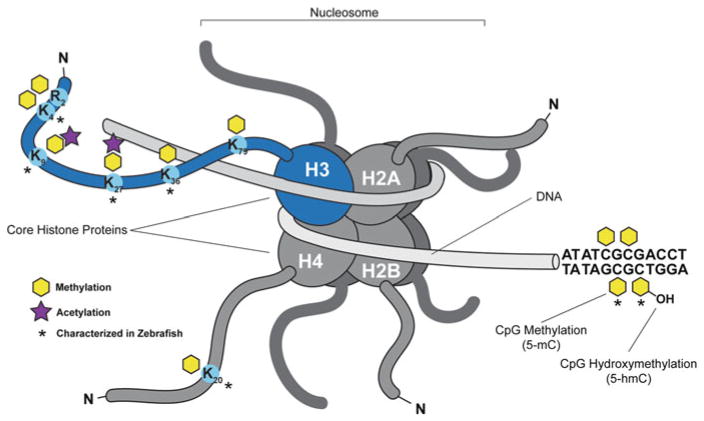
Epigenetic modifications are conserved in zebrafish. Representation of the basic structure of the nucleosome composed of 147 base pairs of DNA wrapped around an octamer of histone H3/ H4 and histone H2A/H2B dimmers. The core histone proteins have long N-terminal tails that extend out from the core particle that is rich in basic amino acid residues, lysine (K) and arginine (R), which can be extensively modified in a reversible, covalent manner. Lysine residues that are mono-, di-, or tri- methylated on histone H3 and H4 that are characterized in zebrafish have been indicated and are conserved across species including H3K4, H3K9, H3K27, H3K36, and H4K20. H3 lysine methylation elicits different transcriptional and structural responses depending on chromatin context and the residues that are modified. Histone acetyl marks have also been indicated and are associated with euchromatic regions amenable to gene transcription. Cytosine residues in DNA can be methylated in a CpG dinucleotide context throughout the genome by DNA methyl-transferases (DNMT), which is a conserved process across vertebrate species and plants. DNA methylation is typically associated with irreversibly silenced regions in heterochromatin. Methylation of DNA can be reversed passively or actively through oxidation of the methyl mark. TET family enzymes carry out active demethylation. Extensive DNA methylation profiling of the zebrafish has been performed in a number of studies