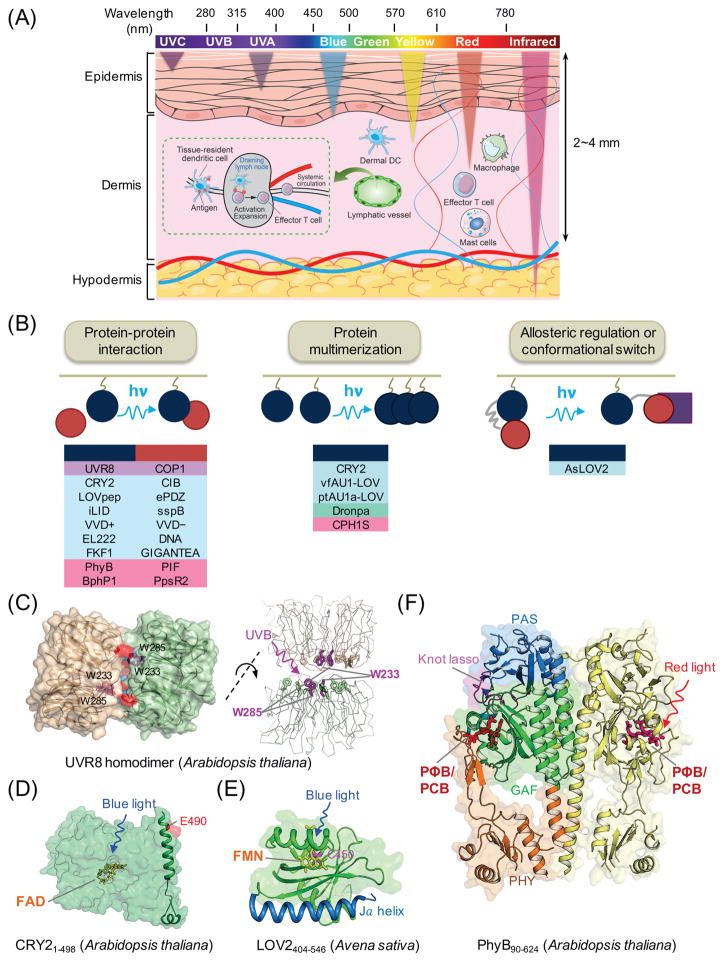
Figure 2. Genetically-encoded photoswitchable modules for optogenetic control of cellular signaling.
(A) Schematic of penetration depth of light emitting at varying wavelengths. Most existing optogenetic tools are activated by blue or yellow light that penetrates up to 0.5-1 mm in depth, with very limited effects on circulating immune cells in living tissues. Tissue-resident dendritic cells (DCs) in the dermis capture antigens (e.g., tumor antigens liberated from melanoma cells) and cross-present them to naïve T cells after migration to draining lymph nodes via lymphatics, leading to activation and expansion of T cells and subsequent trafficking to tumor sites through systemic circulation.
(B) Light-mediated control of cell signaling via photo-manipulation of protein-protein interaction (left), clustering of proteins (middle), or allosteric regulation of protein functions (right). Representative examples of photoresponsive modules are listed under each engineering strategy. UVR8, ultraviolet-B receptor 8; COP1, constitutively photomorphogenic 1; CRY2, cryptochrome-2; CIB, cryptochrome-interacting basic-helix-loop-helix protein; LOV, light-oxygen-voltage; VVD, VIVID protein with a Per-ARNT-Sim domain; FKF1: flavin-binding, kelch repeat, F-box protein; PhyB, phytochrome B; vfAU1/ptAU1, aureochrome 1 from Vaucheria frigida or Phaeodactylum tricornutum.
(C–F) 3D structures of plant-derived photoresponsive domains or proteins that are commonly used in optogenetic applications. The cofactors or chromophores that confer light sensitivities to corresponding proteins are: (C) tryptophan residues for UVR8 (PDB entry: 4DNW); (D) FAD for CRY2 (modeled from CRY1; PDB entry: 1U3D); (E) FMN for LOV2 (PDB entry: 2V0W); and (F) PΦB or PCB for PhyB (PDB entry: 4OUR). FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide; PΦB, phytochromobilin; PCB, phycocyanobilin.