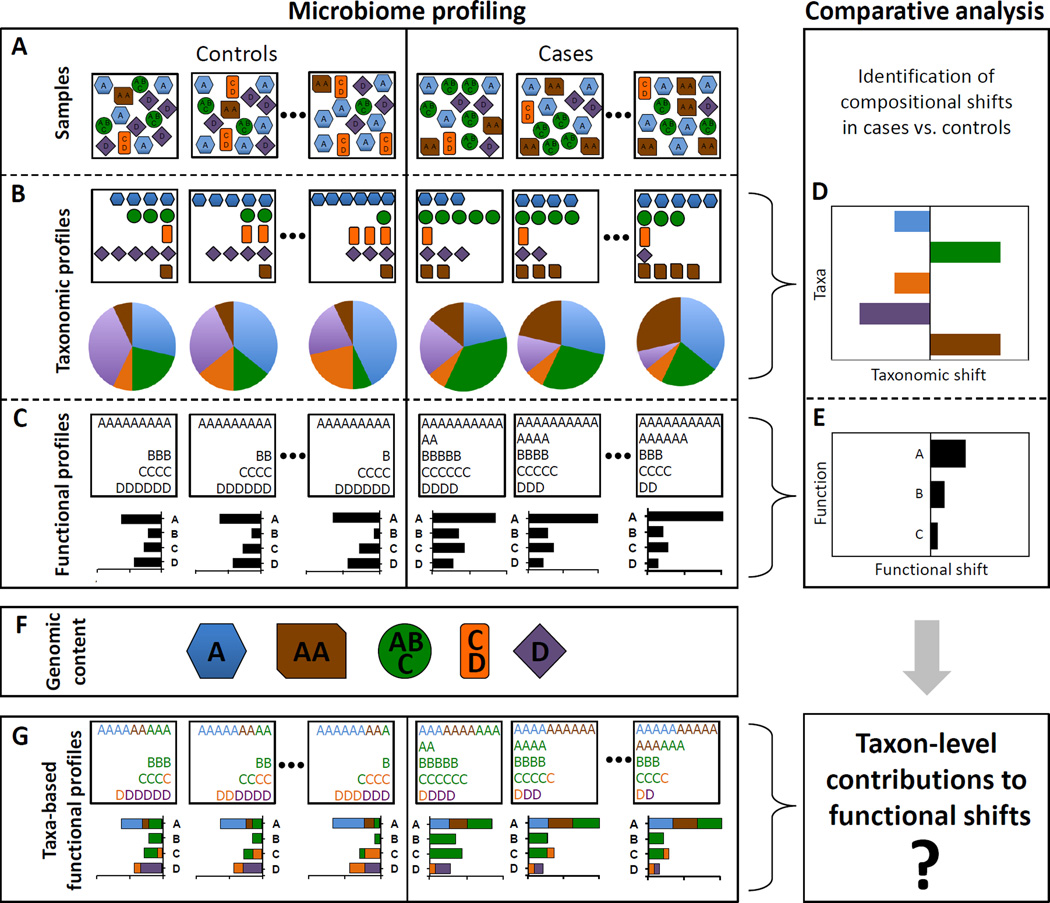
Figure 1. An illustrative example of taxonomic and functional comparative metagenomic analyses.
(A) Community composition of case (right) and control (left) samples. (B) Profiling of the samples’ taxonomic composition, showing the species counts (upper boxes) and relative abundances (pie charts). (C) Profiling of the samples’ functional composition, showing the function counts (upper boxes) and abundances (bar charts). (D–E) Comparative analysis aims to explore compositional shifts in the microbiome, identifying case-associated taxa and/or case-enriched functions. (F) The genomic content of the various community members (denoting the copy number of each gene in their genomes) links the community’s taxonomic and functional profiles. (G) Taxa-based functional profiles provide a mapping from taxonomic to functional compositions, detailing the fraction of each gene’s abundance that originated from each species.