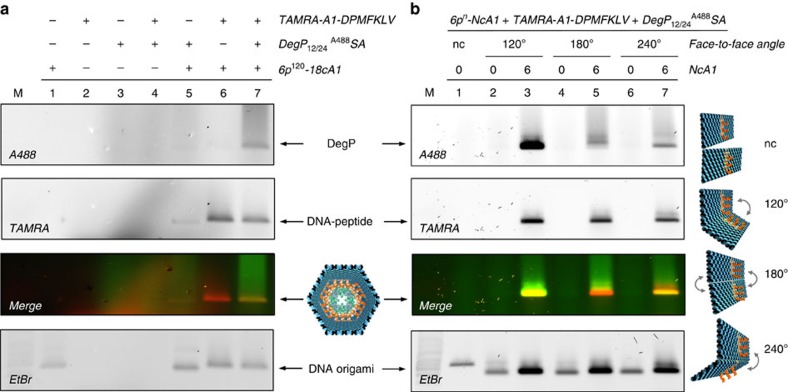
Figure 2. Gel electrophoresis analysis of DegP12/24A488 SA binding to the DNA host.
(a) Analysis of the single components of the complex (lanes 1–3, corresponding, respectively, to the DNA origami host, the TAMRA-labelled DNA–peptide conjugate and the DegP protein) as well as of their mutual interactions (lanes 4–7) indicates specific DNA–protein binding only in presence of the peptide (lane 7). (b) Binding of DegP12/24 to the 6p construct occurs only in presence of PAs (NcA1=6 in lanes 3, 5 and 7) and is dependent on their convergent (120°), randomly oriented (180°) or divergent (240°) arrangement. Only a convergent design of ligands (lane 3) leads to satisfactory yields of binding, whereas undefined (lane 5) or divergent (lane 7) ligand orientation is poorly efficient (yields are in a 8:1.4:1 ratio). As control, a DNA structure lacking the face-to-face connections (nc) has been used (lane 1). Lanes M contained 1 kbp DNA ladder (Roth). The DNA origami structures migrate between the 1.5 and 2.0 kbp bands of the ladder. Gel running conditions: 0.75% agarose in 1 × TBEMg buffer, 4 °C, 3 h at 80 V. Gel imaging was performed with a Typhoon FL900 upon illumination at selected wavelengths to allow detection of the protein (Alexa488), peptide ligand (TAMRA) and DNA (upon ethidium bromide staining). Full gels are shown in Supplementary Figs 34 and 43.