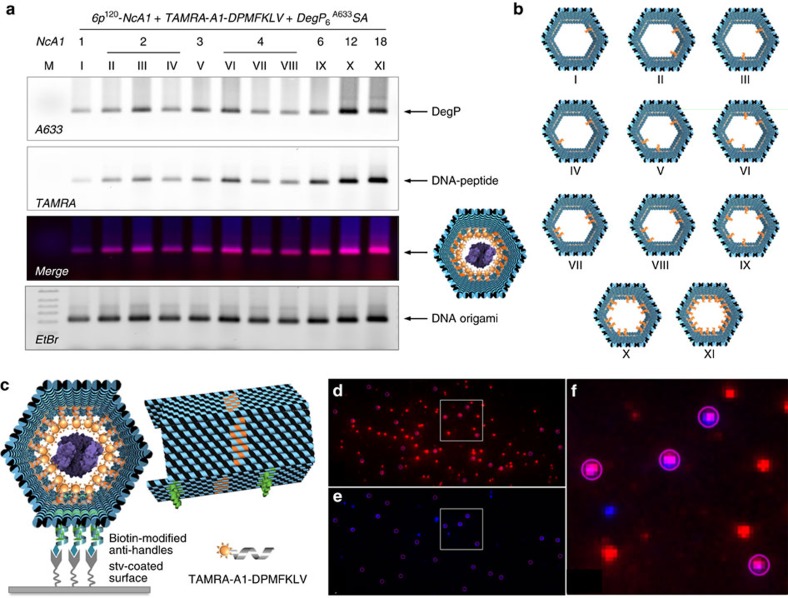
Figure 3. Binding of DegP6A633 SA to diverse 6p120 constructs.
(a) Agarose gel electrophoresis characterization of the binding of DegP6A633SA (labelled at genetically introduced cysteine residues with A633) to diverse 6p120 constructs, differing in the number (NcA1; from 1 to 18) and spatial arrangement of the ligands within the cavity (constructs I to XI, b). The results indicate successful binding for all constructs, with maximal efficiency for a radial distribution of ligands (constructs X and XI). Lane M contained 1 kbp DNA ladder (Roth). The DNA origami structures migrate between the 1.5 and 2.0 kbp bands of the ladder. Gel running conditions: 0.75% agarose in 1 × TBEMg buffer, 4 °C, 3 h at 80 V. Gel imaging was performed with a Typhoon FL900 upon illumination at selected wavelengths to allow detection of the protein (Alexa633), peptide ligand (TAMRA) and DNA (upon ethidium bromide staining). (c) Single-molecule fluorescence characterization of the 6p construct, bearing 18 convergent PAs hybridized with TAMRA-tagged peptide ligands and loaded with a DegP6A647SA protein. Molecules were immobilized on a coverslip surface and were measured using TIRF microscopy. TAMRA (red spots in d) and Alexa647 (blue spots in e) detection channels have been overlapped, indicating clear co-localization of the two species (violet spots showing energy transfer from the donor to the acceptor fluorophore in f, which shows a zoom-in view of the highlighted region in d,e).