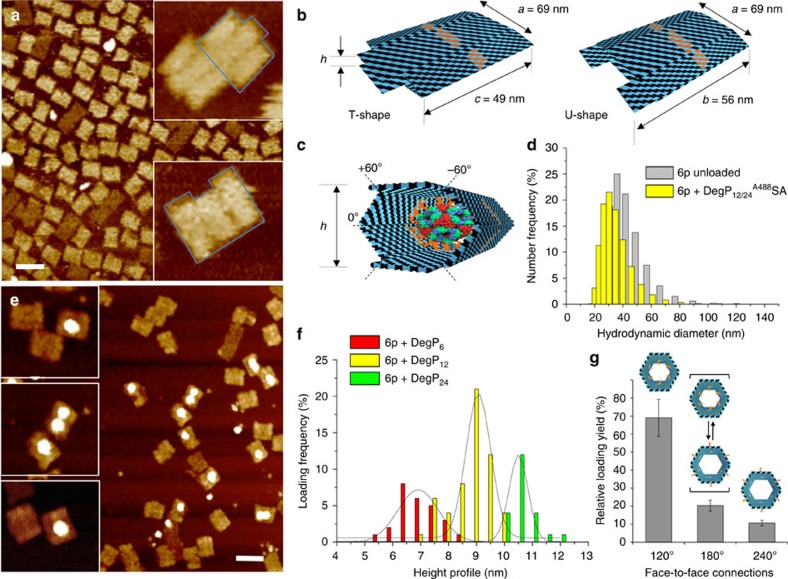
Figure 4. AFM characterization of the unloaded and DegP-loaded host.
(a) The unloaded host deforms during AFM imaging in air, leading to a double-layered structure with one of two possible shapes: either a T or a U shape (b). Whereas the former results from the bending of the structure along the symmetry axis indicated as 0°, the latter is obtained from compression of the structure along the ±60° axes (c). Width and length of the obtained structures are as expected when compressing a correctly formed construct. (d) The hydrodynamic size distribution of the gel-purified DNA origami host either unloaded (grey bars) or loaded (yellow bars) with DegP12 protein was measured by dynamic light scattering, demonstrating correct formation of the hollow structure in solution. (e) Loading of the host with DegP protein mostly results in the appearance of single brighter dots in the centre of the structures, thus indicating successful protein binding in a 1:1 ratio. Scale bars, (a,e) 100 nm. (f) Analysis of the height profile of the structures revealed three distributions centred at ca. 7, 9 and 10.5 nm, corresponding, respectively, to encapsulation of the 6-, 12- and 24-mer (red, yellow and green bars, respectively). Preferential binding selectivity was observed for the 12-mer (more than 50% of the whole population). (g) Systematic analysis of the loading yield revealed most efficient protein encapsulation for a convergent design of multiple ligands (120°). Error bars indicate s.d.'s.