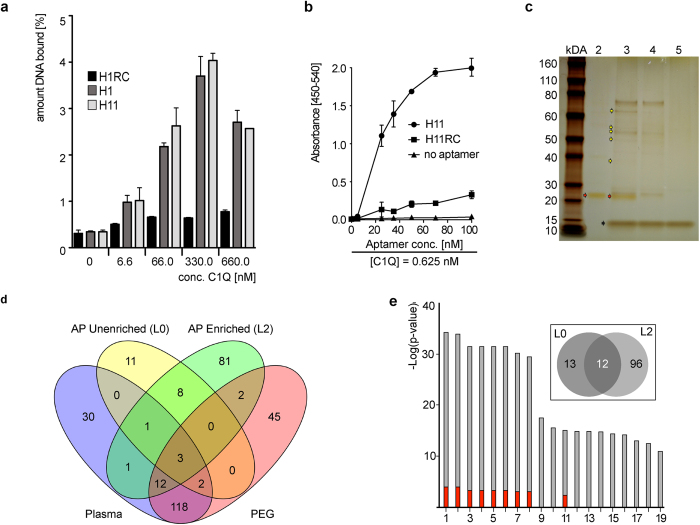
Figure 3. Oligonucleotides identified by ADAPT reveal aptamer-like characteristics.
(a) Filter retention analysis of C1Q-binding by the ssODNs H1 (dark grey columns) and H11 (pale grey columns) at indicated C1Q concentrations. As a control, the reverse complement of H1, H1RC, was used (black columns). (b) ELONA analysis of C1Q-binding by the ssODNs H11 (circles) at indicated concentrations and a fixed C1Q concentration (0.625 nM). As a control, the reverse complement of H11, H11RC, was used (squares). “No aptamer” control (triangles) shows low background binding of detector Streptavidin-HRP. H11 specifically binds C1Q (estimated KD around 40 nM). (c) PAGE analysis of PEG-precipitated and ssODN-associated proteins pulled-down with L2. Lane 1: Molecular weight marker; lane 2: Input library L2; lane 3: Fraction pulled down by biotinylated L2; lane 4: Fraction pulled down by non-biotinylated L2; lane 5: Fraction found in the absence of DNA library; red arrows indicate the ssODN library; yellow arrows indicate protein bands cut out and analysed by LC-MS/MS; black arrows indicate streptavidin monomers leaking from beads. (d) Four-way Venn diagram of proteins detected by LC-MS/MS from Unfractionated plasma, PEG-precipitated plasma, PEG-precipitated plasma in presence of L0 or L2, respectively, purified by streptavidin magnetic beads (background-subtracted; i.e. biotinylated minus non-biotinylated libraries). (e) Gene ontology (GO) cellular component enrichment analysis of the subset of proteins associated with L0 (red bars) and L2 (grey bars) that show a p value of at least 6 × 10−12. Proteins listed are: 1 extracellular region part, 2 extracellular region, 3 extracellular exosome, 4 extracellular membrane-bounded organelle, 5 extracellular organelle, 6 extracellular vesicle, 7 membrane-bounded vesicle, 8 vesicle, 9 organelle, 10 membrane-bounded organelle, 11 extracellular space, 12 focal adhesion, 13 cell-substrate adherence junction, 14 cell-substrate junction, 15 adherence junction, 16 anchoring junction, 17 cellular component, 18 cell junction, 19 blood micro particle. Inset: 108 proteins pulled down by L2 (inset, 96 + 12), cut from the gel shown in (c), and analysed by LC-MS/MS, 13 proteins unique to background-subtracted L0 (inset, 13), and 12 overlapping proteins (inset, 12).