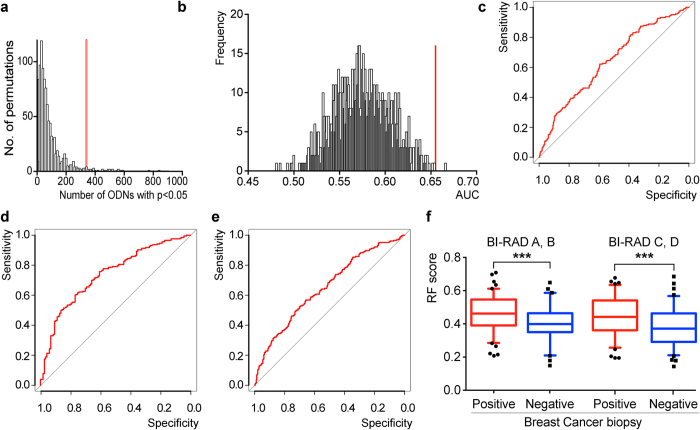
Figure 5. Assessment of ADAPT profiles of 500 clinical samples suggests sample type discrimination.
(a) F-test for cancer versus non-cancer versus negative on original (red line) and permutated (columns) datasets. The number of ssODNs that distinguish breast cancer biopsy positive from negative and self-declared normal samples is significantly higher then can be achieved randomly in 1000 permutations of the original dataset. This shows that the selected ssODNs from L2000 reveal information that distinguishes the analysed cancer population from the other, confirming the applicability of the enrichment strategy and the proper compilation of the small profiling library L2000. (b) OOB AUC from the entire set of 500 clinical samples and permutation analysis of its reliability. AUC in the original dataset is 0.66 (red line), which is significantly higher compared to the majority of 1000 permutations (p = 0.002; columns). (c) ROC curve for cancer versus control samples with AUC = 0.66; p = 0.002. (d) ROC curve for cancer versus healthy donors with AUC = 0.73; p = 0.001. (e) ROC curve for cancer versus biopsy negative with AUC = 0.64; p = 0.024. (f) RF based probability of a cancer incident in both biopsy positive and negative sample types and corresponding radiographic Bi-RAD C&D scores suggesting that ADAPT reliably differentiates cancer from control groups even when using samples from women with dense breast tissue (BI-RAD A&B and C&D Cancer/Control p < 0.001).