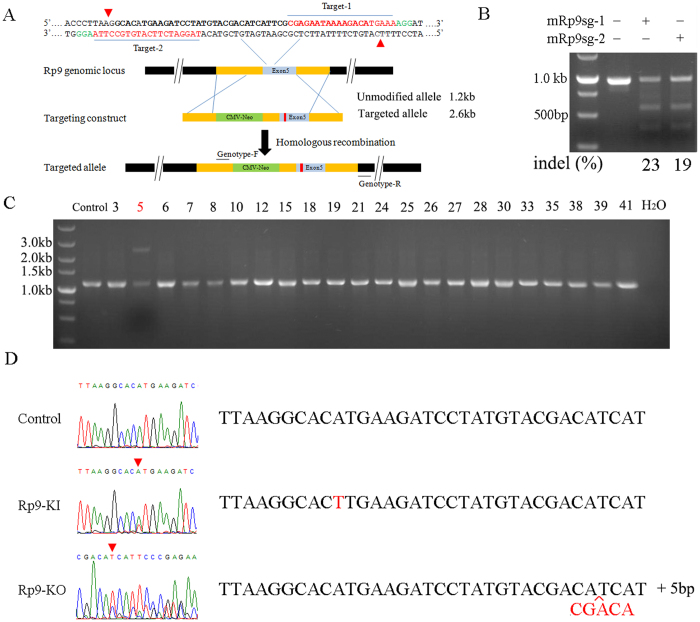
Figure 2. Genome editing via the type II CRISPR system in 661 W cells.
(A) Schematic illustrating Cas9n double nicking the mouse Rp9 locus and strategy of Cas9n pairs- mediated HDR. mRp9sg-1 and mRp9sg-2 sequences are shown in red. Representative cleavage sites are shown by a red triangle for 661 W cells transfected with Cas9n pairs matching target-1 and 2. The targeting vector includes homology arms (HA) flanking a CMV-Neomycin element and Rp9 point mutation. (B) SURVEYOR assay for Cas9 and sgRNA-mediated indels. 661 W cells were transfected by empty vectors or vectors expressing mRp9sg-1 or mRp9sg-2 to test cutting efficiency in the endogenous Rp9 locus. The DNA in the specific cells mentioned above were extracted and performed by PCR and T7EI assays. The percentage of indels was quantified by the ImageJ software. (C) PCR genotyping. As shown, 1 clone (marked with red number) out of 22 randomly selected clones had the expected Rp9 mutation insertion. (D) Representative Sanger sequencing results of the PCR amplicons from 22 clones showing point mutation and insertion (red).