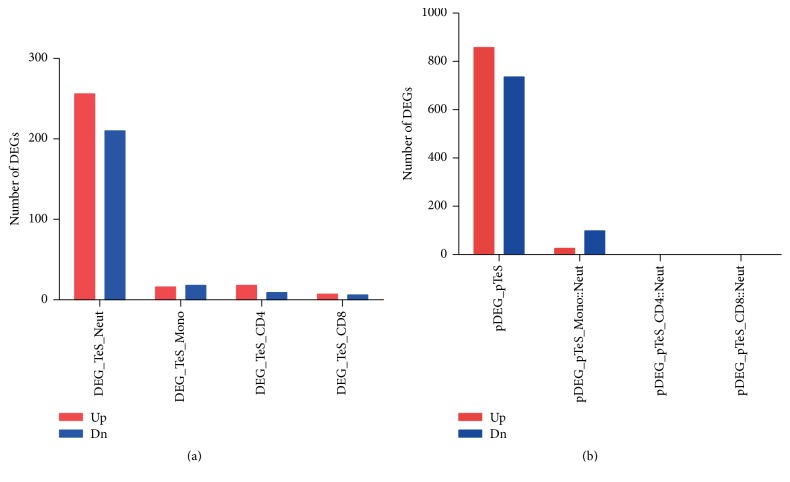
Figure 4.
Number of differentially expressed genes (DEGs) in the four separated cell populations (a) and in putative whole blood transcriptome data (b) of PTB patients. In the normalized transcriptome array data GSE19491 (downloaded from NCBI Gene Expression Omnibus), any probe intensity (probe signal) less than 10 was set to 10. Probes with intensity larger than 10 in at least 50% of the 498 samples were selected. The 16,287 probes so selected were used for the subsequent identification of DEGs in the four separated cell populations as well as in the putative whole blood transcriptome data. Linear Models for Microarray Data (LIMMA) were applied for the identification of DEGs [7]. LIMMA used linear models and empirical Bayes methods (the moderated t-statistic) in assessing differential gene expression. The criteria for DEGs identification were log2 (fold change) ≤ −1 or ≥1 with an adjusted P ≤ 0.01 corrected using Benjamini and Hochberg procedure [7]. Putative whole blood transcriptome data were then generated exactly the same as for the generation of putative PTBsig (pPTBsig) (Figure 1), except that in this case the putative whole blood transcriptome data was from 16,287 probes of gene array whereas pPTBsig had only 393 transcripts/probes. The putative whole blood transcriptome data were normalized based on quantiles for the cross-array comparison and then used for identification of DEGs. DEG_TeS_Neut = DEGs in neutrophils of PTB patients compared to neutrophils of HC donors from Test_set_separated; pDEG_pTeS = putative DEGs of putative whole blood of PTB patients compared to putative whole blood of HC donors based on the four cell populations of Test_set_separated with no exchange of cell proportions; pDEG_pTeS_Mono::Neut = putative DEGs of putative whole blood of PTB patients compared to putative whole blood of HC donors based on the four cell populations with the exchange of cell proportions between monocytes and neutrophils. Other abbreviated names were similarly derived.