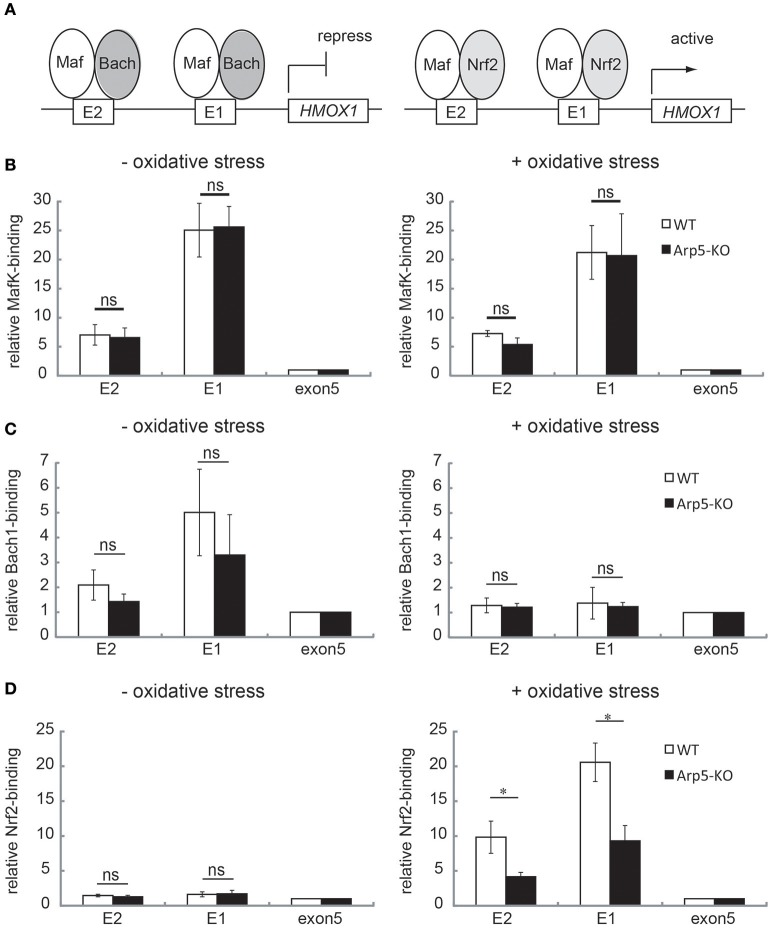
Figure 6.
Analysis of the binding of transcription regulators to the E2 and E1 sites of HMOX1 in wild-type and Arp5-KO cells. (A) In the absence of any oxidative stress (left panel), the repressor Bach1 forms a heterodimer with MafK, which binds to the Maf recognition element (MARE) at the E2 and E1 sites, and represses HMOX1 transcription. In the presence of oxidative stress (right panel), Bach1 is released, and the activator Nrf2 forms a heterodimer with MafK and binds to the MARE elements at E2 and E1. To analyze the binding of these transcriptional regulators, quantitative-ChIP assay was performed using antibodies against MafK (B), Bach1 (C), and Nrf2 (D) in wild-type (open bar) and Arp5-KO (filled bar) cells. The amount of immunoprecipitated fragment was normalized with respect to the input fraction value. Data shown are binding relative to that of exon 5. Averages from at least three independent experiments (± standard deviation) are shown. P-value (Student's t-test) for the difference between WT and Arp5 KO cells is indicated. *P < 0.05.