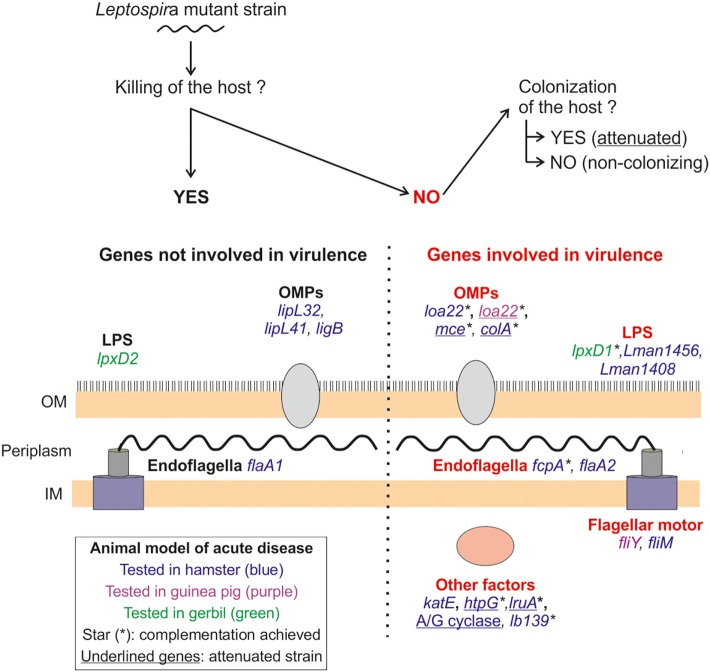
Figure 2.
Leptospira sp. virulence factors identified in animal models of lethal disease. The panel depicts the different factors (in capitals) and genes tested for virulence in different animal models (summarized in Tables 3 and 4) and their localization in Leptospira sp. The mutant strains were inoculated via the intraperitoneal route, using both lethal and sublethal bacterial doses, in different animal models for acute disease: hamster (blue), guinea pig (purple), and gerbil (green). In all cases, the mutant strains were compared with their wild-type counterpart to assess the effect on virulence of the mutated genes. As schematized on top of the panel, we discriminate the genes in two groups; the genes not involved in virulence (on the left side of the panel), since the mutant strain killed the host, and the virulence genes (right side of the panel) determined since the mutant strain did not cause the death of animal. In this category, we further distinguish the non-virulent (NV) mutants that did not colonize target organs and the NV attenuated ones that did. Attenuated mutants are indicated by underlined names. Complementation of mutated genes was not always achieved, and the complemented mutants are indicated with a star (*). OM, outer membrane; IM, inner membrane; OMP, outer membrane protein.