Figure 4.
γH2AX Hotspots in Transformed B Cell Precursors Relate to Transcriptionally Active Genes
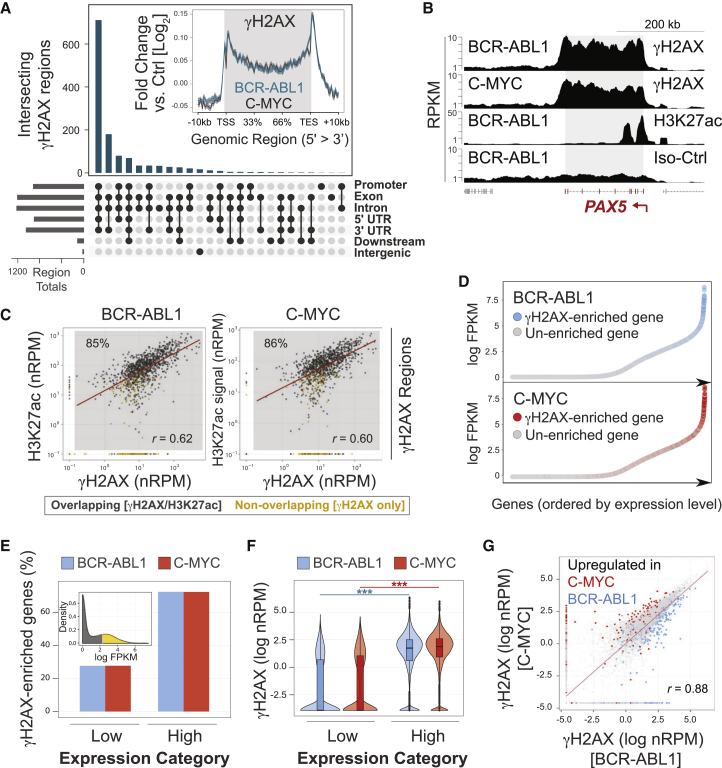
(A) Overlap of γH2AX regions with the indicated gene features for all γH2AX regions identified in transformed B cell precursors. Inset: distribution of γH2AX reads within gene bodies versus upstream and downstream regions for transformed B cell precursors by meta-gene analysis. Signals are normalized to Iso.
(B) A representative custom track of γH2AX versus H3K27ac read densities.
(C) Scatterplots of γH2AX versus H3K27ac intensities for γH2AX regions. Signals are background-subtracted (“normalized”) RPM values for γH2AX and H3K27ac within identified γH2AX regions from the merged γH2AX region list. For better identification of overlapping regions, regions were extended ± 10 kb as described in the Supplemental Experimental Procedures. Linear regression (red line) and Pearson’s correlation (p < 2.2 × 10−16 for both) between γH2AX versus H3K27ac signal intensities is shown for regions that contain both γH2AX and H3K27ac signals (gray shaded area, percentage of total indicated). See Figure S4B for meta-gene analysis of H3K27ac.
(D) Relative gene expression levels ranked by fragments per kilobase of transcript per million reads (FPKM) for BCR-ABL1- and C-MYC-transformed B cell precursors. Genes overlapping γH2AX-enriched regions are highlighted as indicated.
(E and F) Genes were categorized as high- or low-expressed relative to the median expression level across the population of all expressed genes. Inset, left: kernel density plot of FPKM values; colors indicate expression categories. Relative distribution of γH2AX-enriched genes between expression categories (E) and distribution of normalized γH2AX values measured for extended gene regions between expression categories (F) are shown. The shaded area indicates kernel density overlaid with a standard boxplot, and stars indicate the highly significant difference between categories (p < 1 × 10−15; Mann-Whitney U test).
(G) Scatterplot of the difference in γH2AX signal (nRPM) across extended gene regions between BCR-ABL1- and C-MYC-transformed B cell precursors. Differentially expressed genes are highlighted in blue or red as indicated. Linear regression (red line) and Pearson’s rho (r) are indicated.