Figure 5.
Analysis of Convergent and Divergent Transcription, R Loop-Forming Sequences, and Early-Replicating Fragile Sites at γH2AX Regions
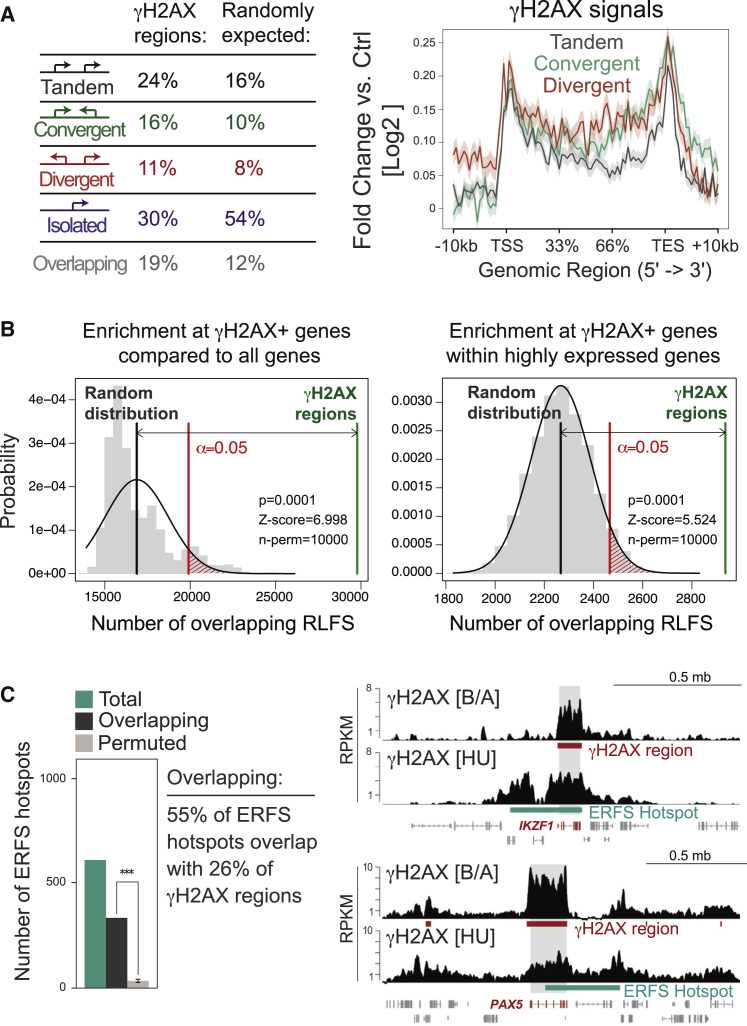
(A) Left: gene pairs within 10 kb were categorized as tandem, convergently, or divergently transcribed, as indicated in the respective diagrams, or as isolated (no additional gene within 10 kb) or overlapping. The proportion of genes in each subset is indicated for both γH2AX region-associated genes (left) and the distribution across the whole genome (right). Right: average fold change in γH2AX signal in BCR-ABL1 versus isotype control across gene bodies for each transcription orientation category, normalized for gene lengths (p < 1 × 10−6 for both convergent versus tandem and divergent versus tandem comparisons using Mann-Whitney test).
(B) Analysis for the enrichment of predicted RLFSs at γH2AX-associated genes. The total count of RLFSs overlapping γH2AX-associated genes (green line) was compared with the permuted null distribution of total RLFSs (gray bars) measured by random sampling of the gene population (the black line indicates mean, and red indicates a significance threshold of 0.05). Significant enrichment of RLFSs at γH2AX-associated genes is observed both among all genes (left) and when analysis is restricted to only the 1,000 most highly expressed genes (right).
(C) Left: bar diagram showing the number of ERFS hotspots (Total) versus the number of these sites that overlap with γH2AX regions in transformed B cell precursors (Overlapping). The significance is indicated by comparison with the mean permuted number of overlaps from random re-sampling of equivalently sized regions within the genome. Right: HU-induced versus BCR-ABL1-induced γH2AX signal patterns at two representative loci by custom track visualization. The bars indicate the ERFS hotspots (mint) and BCR-ABL1-induced γH2AX regions (red). Grey boxes indicate gene body location of the highlighted gene.