Abstract
Synonymous variations in protein-coding sequences alter protein expression dynamics, which has important implications for cellular physiology and evolutionary fitness, but disentangling the underlying molecular mechanisms remains challenging.
The encoding of twenty amino acids by 61 codons enables a vast number of synonymous gene sequences to express the same protein. A fundamental question in molecular biology concerns how this redundancy in the genetic code is exploited physiologically and evolutionarily. However, despite the proximity of this question to the heart of the Central Dogma, significant uncertainty remains concerning the answers. Two recently published papers (Kelsic et al. 2016, Frumkin et al. 2017) illustrate the power of high-throughput systems biology methods to provide insight into complex biological questions of this kind, while they simultaneously raise new questions highlighting the complexity of the biological and evolutionary phenomena related to codon usage.
Both of these papers used deep sequencing methods to characterize the relative growth rates or comparative “fitness” of E. coli cells in mixed populations expressing different genetic variants of a single protein molecule. Kelsic et al. used multiplex automated genome engineering (MAGE) to mutate every codon in the chromosomal copy of the essential infA gene, which encodes translation initiation factor 1 (IF1). Fitness in this experiment monitors the effect of each genetic variation on net cellular IF1 activity, which presumably correlates both with the functional competency and the expression level of this abundant protein that is required for translation initiation. Their central conclusion is that variations in synonymous codon usage have the strongest effects at the beginning or “head” of the coding sequence, where they act by altering mRNA folding properties that modulate translation initiation rate. This conclusion echoes that of several recent studies (Goodman et al., 2013; Boel et al., 2016), but the comprehensive and compelling nature of the data they obtained from a single, technically coherent experiment illustrates the impressive power of high-throughput systems biology approaches. Their data furthermore show that a stem-loop structure upstream of the ribosome-docking site promotes greater fitness, presumably by increasing the efficiency of translation initiation, consistent with previous studies (Espah Borujeni et al., 2014) that included a putative "ribosome standby site" in this region of some E. coli mRNAs. The authors conclude that deviations from the folding properties of the native mRNA sequence generally tend to reduce fitness and presumably the expression level of this protein. However, the Kelsic et al. data also show some features that are inconsistent with trends reported in prior literature, highlighting the mechanistic and evolutionary complexities that can produce seemingly idiosyncratic codon-usage effects.
Frumkin et al. have re-analyzed a library of 14,000 genes with sequence variations in the regions controlling transcription and translation initiation of a green fluorescent protein (GFP) reporter (Goodman et al., 2013). The sequence variations in this library occur in an 11-residue amino acid extension appended at the N terminus of GFP, a design intended to minimize the influence of mRNA sequence variations in the “head” of the coding sequence on GFP folding and fluorescence. The initial application of this library focused on the influence of the sequence variations on cellular GFP fluorescence level, and this analysis pointed toward the same conclusion emphasized by Kelsic et al., i.e., that variations in synonymous codon usage in the head modulate translation initiation rate by changing mRNA folding properties in this region. Frumkin et al. extended the characterization of this library by using deep sequencing to evaluate the competitive fitness of cells expressing every variant in a mixed population that was serially re-cultured in Luria Broth (LB) every day for 12 days. They dubbed this method, which is similar to that employed by Kelsic et al., “FitSeq”. They conclude that pressure to conserve metabolic resources is responsible for two trends observed in their fitness dataset. The first is enrichment in cells containing sequences that promote translation of many protein molecules from a single mRNA molecule, which reduces the metabolic burden associated with transcription. The second is depletion of cells containing the sequences encoding hydrophobic amino acids in the N-terminal extension on GFP, presumably because their inclusion increases the metabolic burden associated with amino acid synthesis. These conclusions highlight an additional complexity potentially influencing the biology of synonymous codon usage, beyond those related to the mechanistic complexities of translation initiation and transcription/translation coupling (Figure 1). If evolutionary optimization of mRNA sequence and codon usage is influenced by metabolic factors, it will depend on the environmental niches and evolutionary history of an organism, factors that may not be replicated or even accessible under laboratory conditions. This point is clearly conceptually important, but some of the results and interpretations reported by Frumkin et al. raise significant questions.
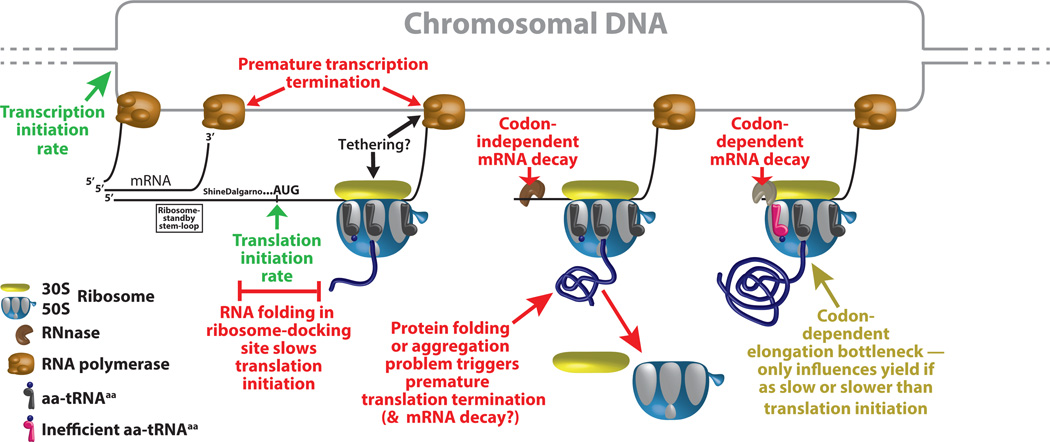
Figure 1. Schematic diagram of mechanistic processes influencing protein expression level.
Processes promoting higher expression are labeled in green, while those that attenuate expression are labelled in red. Chromosomal DNA is shown and labeled in gray, RNA transcripts in black, and nascent protein in navy blue. The co-transcriptional translation schematized here only occurs in prokaryotes, but all the same molecular mechanisms likely influence protein expression in eukaryotes with the exception of those involving direct transcription-translation coupling. The Shine-Dalgarno and ribosome standby-site stem-loop (Borujeni et al., 2014) contribute to control of translation initiation in prokaryotes, while other factors control this process in eukaryotes.
These new high-throughput fitness studies combined with other recent large-scale studies employing different methods (Goodman et al., 2013; Boël et al, 2016) create a broad consensus that codon usage at the beginning of the coding sequence of a protein is constrained by optimization of mRNA folding. This observation accounts for the longstanding observation that codons that are used infrequently elsewhere in a given genome are often enriched at the start of protein-coding sequences (Chen et al., 1990). However, the factors determining codon frequency and its relationship to translation efficiency remain controversial.
Historical biochemical and molecular biological literature tracing back five decades claims that infrequently used codons tend to be translated inefficiently and reduce protein expression level (Chen and Inouye, 1990), due to a kinetic barrier caused by the low concentration of the cognate tRNAs (Caskey et al., 1968; Tuller et al., 2010; Yu et al., 2015). In vivo studies contributing to this literature generally examined protein overexpression (Chen and Inouye, 1990), which can create an imbalance between the cellular tRNA pool and the codon content of the translating mRNA pool (Caskey et al., 1968; Tuller et al., 2010), parameters that are generally closely balanced under physiological conditions. Recent ribosome profiling studies (Mohammad et al., 2016) and large-scale protein-expression studies (Boël et al, 2016) have raised doubts about both the generality of the underlying mechanistic model and the premise that infrequently used codons impede protein translation. These doubts are reinforced by the synonymous codon-influence effects observed by Kelsic et al. outside the head region of the gene encoding IF1 because these show a low correlation with codon frequency that is borderline in statistical significance after correction for multiple-hypothesis testing. A similarly weak correlation was observed in a recently published large-scale protein overexpression study (Boël et al, 2016), which generated a new codon-influence metric that correlates significantly with mRNA lifetimes under physiological conditions in E. coli. This study echoed other recent studies showing similar correlations between codon usage and mRNA decay rates in the yeast Saccharomyces cerevisiae (Presnyak et al, 2015) and the zebrafish Danio rerio. These parallel results linking codon usage to mRNA decay rate in organisms from three widely separated branches on the tree of life have opened an important new chapter on the biology and biochemistry of synonymous codon usage.
Notably, there are differences in the specific codon effects Kelsic et al. observed in gene encoding IF1 and those observed in the large-scale study that revealed the connection between codon usage and mRNA decay in E coli., which examined over-expression of 6,348 different proteins (Boël et al, 2016). Although synonymous substitutions at some positions outside the head region of IF1 produce substantial changes in fitness, very few individual codons show a consistent and statistically significant influence in this region of the gene. The infA codons that appear to be beneficial seem biased towards having a C base in the wobble position, an effect that has not appeared in prior codon-influence metrics. Furthermore, the ATA codon for isoleucine and the CGG codon for arginine, which have been identified consistently in previous studies as tending to reduce expression, do not reduce fitness in their study. The apparently idiosyncratic nature of the codon effects observed in the impressively comprehensive study by Kelsic et al. highlights the challenges that remain relative to understanding the physiological and evolutionary implications of codon usage.
These challenges arise from the confluence of many mechanistic and evolutionary factors that can influence variations in synonymous codon usage and their effects on protein expression (Figure 1). Net protein expression level depends in a straightforward way on the concentration of the corresponding mRNA and the rate of translation initiation on that mRNA. Variations in codon usage near the translational start codon have long been known to influence translation initiation rate, because the mRNA in that region must be accessible in a single-stranded conformation for docking of the ribosome and formation of the ribosomal initiation complex (IC), and both formation of stable RNA 2° structures and the transient hydrogen-bonding propensity of the nucleotide bases in this region can inhibit docking and IC formation (Borujeni et al., 2014; Goodman et al., 2013; Boël et al, 2016). Modulation of alternative mRNA 2° structures in this region is known to be exploited for translational regulation of protein expression (Nudler and Gottesman, 2002) and may contribute to one idiosyncrasy observed by Kelsic et al., a somewhat beneficial effect on fitness of base-pairing between the first four codons in IF1 and downstream regions of its coding sequence. Previous large-scale studies suggest that base-pairing in the first six codons generally tends to attenuate protein expression (Boël et al., 2016), so the contrary trend observed by Kelsic et al. may reflect an uncharacterized regulatory mechanism in which formation of alternative mRNA 2° structures in this critical region control the initiation of IF1 translation. The mRNA sequence and 2° structures in the ribosome-docking region can furthermore influence premature transcription termination, which in prokaryotes can be directly modulated by the efficiency of translation initiation (Figure 1), creating potentially complex regulatory circuitry (Nudler and Gottesman, 2002). These mechanisms influencing the effective transcription rate have the potential to couple mRNA concentration to codon usage near the translational start codon. The confluence of so many mechanistic factors explains how synonymous sequence variations in this region can have idiosyncratic effects depending on the regulatory physiology of individual genes.
As explained above, recent results from E. coli, yeast, and zebrafish have shown that variations in codon usage throughout a gene can also influence mRNA decay rate (Presnyak et al, 2015; Boël et al., 2016)), presumably via the action of endoribonucleases allosterically controlled by the decoding process on the ribosome. Coupling to mRNA decay represents another mechanism by which variations in synonymous codon usage can influence both mRNA and protein expression levels. These recent studies and others have raised questions about the generality and physiological importance of codon influence on translation elongation rate, which can only influence protein expression level when the elongation rate is reduced to be comparable to or slower than the translation initiation rate (Figure 1). The clearly demonstrated ability of mRNA sequence variations near the translational start codon to alter translation initiation rate implies that synonymous codon variations in that region of the gene could change the influence of synonymous codon variations downstream in the gene. Yet another confounding factor is that co-translational folding vs. aggregation of the protein being synthesized on a ribosome may influence elongation rate as well as mRNA decay rate (Figure 1), while the protein folding efficiency itself can be influenced by the local translational elongation rate (Yu et al., 2015).
Beyond these myriad mechanistic complexities related to transcription/translation dynamics, the fitness dataset generated by Frumkin et al. shows several effects that could be attributable to stress on cellular metabolic resources caused by mRNA transcription and protein expression. One noteworthy feature in their dataset is that higher expression of GFP generally reduces cellular growth rate and fitness. This observation has sobering implications regarding the widespread use of GFP to study the mechanism and physiology of protein expression (e.g., Goodman et al., 2013), because it suggests genetic selection and segregation in response to the stress could influence results. While the effects observed by Frumkin et al. could reflect metabolic stress, they could alternatively derive from toxicity caused by adventitious molecular interactions of the GFP protein itself or from stress associated with the protein-folding process. Although Frumkin et al. used a variant of GFP that has been engineered to fold more efficiently, its folding in vivo in E. coli could still be problematic and impaired by the hydrophobic N-terminal extensions they observe to reduce fitness. Many studies demonstrate that protein folding can be coupled to translation, and there is evidence that folding problems, which can clearly create stress, can influence translation efficiency and mRNA decay (Figure 1). The proposal of Frumkin et al. that hydrophobic extensions reduce fitness because of their greater metabolic cost suggests that their deleterious influence should be enhanced in cells growing in minimal medium compared to the amino-acid-rich LB medium used in their study. Additional experimentation of this kind could help discriminate metabolic effects from effects related to the influence of protein folding on transcriptional and translational dynamics (Figure 1).
The recently published papers by Kelsic et al. (2016) and Frumkin et al. (2017) both illustrate the impressive power of systems biology approaches to rapidly characterize biological systems in greater depth and breadth than possible using traditional genetic and biochemical experimentation. However, the conceptual, mechanistic, and evolutionary complexities influencing codon usage are likely to keep both systems biologists and traditional molecular biologists busy for a considerable period of time before a confident understanding is achieved of the related principles and phenomena.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Boel G, Letso R, Neely H, Price WN, Wong K-H, Su M, Luff JD, Valecha M, Everett JK, Acton TB, et al. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature. 2016;529:358–363. doi: 10.1038/nature16509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borujeni AE, Channarasappa AS, Salis HM. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 2014;42:2646–2659. doi: 10.1093/nar/gkt1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey CT, Beaudet A, Nirenberg M. RNA codons and protein synthesis. 15. Dissimilar responses of mammalian and bacterial transfer RNA fractions to messenger RNA codons. J. Mol. Biol. 1968;37:99–118. doi: 10.1016/0022-2836(68)90076-4. [DOI] [PubMed] [Google Scholar]
- Chen GF, Inouye M. Suppression of the negative effect of minor arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucl. Acids Res. 1990;18:1465–1473. doi: 10.1093/nar/18.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frumkin Schirman, et al. Gene architectures that minimize cost of gene expression. Mol. Cell. 2017;65:142–153. doi: 10.1016/j.molcel.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman DB, Church GM, Kosuri S. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013;342:475–479. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- Kelsic Chung, et al. RNA structural determinants of optimal codons revealed by MAGE-seq. Cell Systems. 2016;3:563–571. doi: 10.1016/j.cels.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Woolstenhulme CJ, Green R, Buskirk AR. Clarifying the Translational Pausing Landscape in Bacteria by Ribosome Profiling. Cell Rep. 2016;14:686–694. doi: 10.1016/j.celrep.2015.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli. Genes to Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen Y-H, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, et al. Codon Optimality Is a Major Determinant of mRNA Stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An Evolutionarily Conserved Mechanism for Controlling the Efficiency of Protein Translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Yu CH, Dang Y, Zhou Z, Zhao F, Sachs M, Liu Y. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell. 2015;134:341–352. doi: 10.1016/j.molcel.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]