Abstract
Small ubiquitin-like modifier (SUMO) conjugation (SUMOylation) plays key roles in neurologic function in health and disease. Neuronal SUMOylation is essential for emotionality and cognition, and this pathway is dramatically activated in post-ischemic neurons, a neuroprotective response to ischemia. It is also known from cell culture studies that SUMOylation modulates gene expression. However, it remains unknown how SUMOylation regulates neuronal gene expression in vivo, in the physiologic state and after ischemia, and modulates post-ischemic recovery of neurologic function. To address these important questions, we used a SUMO1-3 knockdown (SUMO-KD) mouse in which a Thy-1 promoter drives expression of 3 distinct microRNAs against SUMO1-3 to silence SUMO expression specifically in neurons. Wild-type and SUMO-KD mice were subjected to transient forebrain ischemia. Microarray analysis was performed in hippocampal CA1 samples, and neurologic function was evaluated. SUMOylation had opposite effects on neuronal gene expression before and after ischemia. In the physiological state, most genes regulated by SUMOylation were up-regulated in SUMO-KD compared to wild-type mice. Brain ischemia/reperfusion significantly modulated the expression levels of more than 400 genes in wild-type mice, with a majority of those genes upregulated. The extent of this post-ischemic transcriptome change was suppressed in SUMO-KD mice. Moreover, SUMO-KD mice exhibited significantly worse functional outcome. This suggests that suppression of global gene expression response in post-ischemic brain due to SUMO knockdown has a negative effect on post-ischemic neurologic function. Together, our data provide a basis for future studies to mechanistically link SUMOylation to neurologic function in health and disease.
Keywords: brain ischemia, SUMO, microarray, knockdown, transgenic mice
INTRODUCTION
Small ubiquitin-like modifier (SUMO) conjugation (SUMOylation) is a post-translational protein modification whereby SUMOs are covalently conjugated to lysine residues in target proteins (Flotho and Melchior, 2013). This is an energy-dependent process that is regulated via the action of activating (E1, ATP-dependent), conjugating (E2), and ligating (E3) enzymes. In mammalian cells, three SUMO isoforms have been identified: SUMO1, SUMO2, and SUMO3. SUMO2 and SUMO3 are highly homologous, and are usually referred to as SUMO2/3. SUMO1, however, shares only about 50% homology with SUMO2/3. Notably, while SUMO1 knockout mice and SUMO3 knockout mice do not show any obvious abnormality, SUMO2 knockout is lethal in the embryonic stage (Evdokimov et al., 2008; Wang et al., 2014b).
SUMOylation can modify the activity, stability, and function of target proteins, and thereby modulate almost all major cellular pathways (Flotho and Melchior, 2013). Our knowledge about the significance of SUMO conjugation in cellular homeostasis is primarily based on results from cell culture experiments. In the physiologic state, most SUMO1 is conjugated to target proteins, predominately the Ran GTPase-activating protein-1 (RanGAP1). Thus, under stress conditions, there is only small change in SUMO1 modification profiles. In contrast, most SUMO2/3 is present as free SUMO in the physiologic state. However, under a variety of stress conditions, including heat, metabolic, and oxidative stress, SUMO2/3 conjugation is dramatically activated (Saitoh and Hinchey, 2000; Yang et al., 2012).
Many of the SUMO targets are nuclear proteins involved in gene expression (Golebiowski et al., 2009; Yang et al., 2014). It has been reported that SUMOylation can both activate and suppress transcription. In most cases, however, gene expression is suppressed when the respective transcription factor is SUMO-conjugated (Gill, 2005; Chymkowitch et al., 2015). The mechanisms that link SUMO conjugation to gene expression are still not fully understood. Generally, experimental studies have focused on individual transcription factors to clarify the role of SUMOylation in expression of target genes. Recently, chromatin immunoprecipitation coupled with next-generation sequencing (ChIP-seq) was used to more completely define these mechanisms (Liu et al., 2012; Neyret-Kahn et al., 2013; Seifert et al., 2015). These studies were carried out in cell cultures. A genome-wide analysis of the effects of SUMOylation on gene expression in vivo has not yet been performed.
SUMO conjugation plays a pivotal role in neurologic function in the physiologic and pathologic state. For example, SUMOylation is essential for emotionality and cognition, and silencing SUMO1-3 expression specifically in neurons impairs episodic and fear memories (Wang et al., 2014a). However, the pathways that link SUMOylation to memory processes have not been identified. Furthermore, SUMOylation is associated with brain ischemia/stroke and degenerative diseases (Yang et al., 2008b,c; Flotho and Melchior, 2013; Krumova and Weishaupt, 2013). Transient brain ischemia is a severe form of metabolic stress that triggers dramatic activation of SUMO2/3 conjugation, and to a lesser extent, SUMO1 conjugation (Yang et al., 2014). It has been proposed that this is a protective response that shields neurons from damage induced by ischemia (Yang et al., 2008a, 2016; Lee and Hallenbeck, 2013). Indeed, results from in vitro and in vivo studies support this notion. For example, neurons in which SUMO2/3 expression is silenced by lentiviral delivery of SUMO2 and SUMO3 microRNAs (miRNAs), are highly sensitive to transient oxygen/glucose deprivation (OGD, an experimental model that simulates ischemia in cells), whereas transgenic mice overexpressing SUMO conjugating enzyme Ubc9, have higher levels of SUMO1- and SUMO2/3-conjugated proteins and smaller infarcts after stroke (Datwyler et al., 2011; Lee et al., 2011). However, we still do not know the role of SUMO conjugation in post-ischemic neurologic function, which ultimately defines quality of life for patients suffering from ischemic brain damage, and how SUMOylation modulates the genome regulated by transient ischemia. Here, we report our findings from the first experimental study that clarifies the contribution of SUMO conjugation to pre- and post-ischemic gene expression and functional outcome. For this study, we used a recently developed neuron-specific SUMO1-3 knockdown (SUMO-KD) mouse model (Wang et al., 2014a).
EXPERIMENTAL PROCEDURES
Animals
All animal experiments were approved by the Duke University Animal Care and Use Committee. A total of 72 mice were used in this study. SUMO-KD transgenic mice were previously generated in our laboratory (Wang et al., 2014a). In this transgenic mouse model, the transgene contains 3 distinct miRNAs that target SUMO1, 2, and 3, and are expressed under the control of the neuron-specific Thy1 promoter. Green fluorescent protein (GFP) is co-expressed as indicator of transgene expression. SUMOKD mice have been backcrossed with C57Bl/6 mice for more than 10 generations. Male SUMO-KD and wild-type littermates (2–3 months old) were used in this study.
Transient forebrain ischemia
Transient forebrain ischemia was performed as described previously (Yang et al., 2008c). Briefly, anesthesia was induced with 5% isoflurane and maintained with 1.5–1.8% isoflurane during surgery. The rectal temperature was maintained at 37.0 °C ± 0.2 °C by surface heating or cooling. PE-10 tubes were inserted into the right femoral artery and the right internal jugular vein to continuously monitor arterial blood pressure and to withdraw blood, respectively. Forebrain ischemia was induced by a combination of 10-min bilateral common carotid artery occlusion, and blood withdrawal-induced hypotension (mean arterial blood pressure = 30 mmHg). To end the ischemic episode, the carotid arteries were de-occluded and withdrawn blood was re-infused. Sham-operated mice underwent the same procedures except carotid artery occlusion and blood withdrawal. To determine whether SUMO knockdown had any effect on blood flow reduction in our transient forebrain ischemia model, a cohort of mice was subjected to blood flow measurements. Before inducing ischemia, a microprobe (Moor) was affixed to the surface of the right parietal skull to monitor regional cerebral blood flow (rCBF) in the middle cerebral artery territory by Laser Doppler flowmetry (Moor).
Tissue sample preparation
At the indicated times of reperfusion, mice were sacrificed, and brains were quickly removed. CA1 and cortex samples were excised in a cryostat set at −20 °C. Tissue samples were stored at −80 °C and later used for RNA or protein preparation. For immunohistochemistry analysis, transcardial perfusion was performed using 4% paraformaldehyde. Brains were collected and fixed overnight. The fixed brains were either embedded in paraffin or immersed in 20% sucrose at 4 °C before stored in −80 °C.
Microarray analysis
CA1 subfield tissue samples were harvested from 4 experimental groups: wild-type sham (WS), wild-type ischemia (WI), SUMO-KD sham (TS), and SUMO-KD ischemia (TI). Post-ischemia samples were collected at 3 h reperfusion. For each group, samples were prepared in triplicate. To minimize variation in biological replicates, CA1 subfield samples from two mouse brains were pooled, and used to prepare total RNA for one independent microarray sample. The Affymetrix GeneChip Mouse Genome 430A 2.0 Array, which contains approximately 14,000 well characterized mouse genes, was used. Synthesis of cDNA, labeling of samples, and array processing was performed at the Duke Sequencing and Genomic Technologies facility (Yang et al., 2009). Partek Genomics Suite 6.6 (Partek) was used to identify differentially expressed genes, and to perform principal component analysis (PCA). Robust multi-chip analysis (RMA) normalization was performed on the entire data set. The differentially expressed genes were selected based on a p value <0.05 (as determined by ANOVA), and a fold-change ≥2. The differentially expressed genes were further analyzed using PANTHER (http://www.pantherdb.org/) and DAVID (https://david.ncifcrf.gov/) to identify enriched genes according to gene ontology (GO) classifications and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Quantitative PCR
Quantitative real-time PCR (qPCR) was performed as previously described (Yang et al., 2009). In short, total RNA was prepared from CA1 tissue samples using the TRIzol reagent (Invitrogen). Total RNA (200 ng) was reverse transcribed into cDNA (Invitrogen). qPCR was performed in a Lightcycler 2.0 (Roche). Fold changes were calculated according to the comparative threshold cycle (Ct) method (Schmittgen and Livak, 2008). All primers used in this study are listed in Table 1.
Table 1.
List of primers
| Gene | Primer sequences (5′->3′) |
|---|---|
| Sumo1 | Forward: CAGGAGGCAAAACCTTCAAC |
| Reverse: CTCCATTCCCAGTTCTTTCG | |
| Sumo2 | Forward: ACGAGAAACCCAAGGAAGGA |
| Reverse: CTCCATTTCCAACTGTGCAG | |
| Sumo3 | Forward: AGAAGCCCAAGGAGGGTGT |
| Reverse: CCTCGGGAGGCTGATCCT | |
| Atf3 | Forward: CCAGGTCTCTGCCTCAGAAG |
| Reverse: CCTTCAGCTCAGCATTCACA | |
| Hspb1 | Forward: GCCTCTTCGATCAAGCTTTC |
| Reverse: CCTCAGGGGATAGGGAAGAG | |
| Fos | Forward: ATGGGCTCTCCTGTCAACAC |
| Reverse: GCAGCCATCTTATTCCGTTC | |
| Arc | Forward: AGCAGCAGACCTGACATCCT |
| Reverse: GTGATGCCCTTTCCAGACAT | |
| Rgs2 | Forward: GAGGAGAAGCGGGAGAAAAT |
| Reverse: GAGGACAGTTTTTGGGGTGA | |
| Tcfl5 | Forward: GGAGGACCGCTTCAACAGTA |
| Reverse: GGCAGTCCAATATCCTGGTG | |
| Cxcl12 | Forward: GCGCTCTGCATCAGTGAC |
| Reverse: TAATTTCGGGTCAATGCACA | |
| Socs2 | Forward: CGCGAGCTCAGTCAAACAG |
| Reverse: GCAGAGTGGGTGCTGATGTA | |
| Ogt | Forward: GCGTTTTCCAGCAGTAGGAG |
| Reverse: CCAGACTTTGCCACGAATTT | |
| Slco1a4 | Forward: AATGCCAAAGAGGAGAAGCA |
| Reverse: TGGGAAATTGTCACAGGTCA | |
| Rasl10a | Forward: TCATCCGCCAATTCTTGTTT |
| Reverse: TTGTTGCCTACCACCAGGAT | |
| β-actin | Forward: TAGGCACCAGGGTGTGATG |
| Reverse: GGGGTGTTGAAGGTCTCAAA |
Western blot analysis
Western blotting was conducted as described before (Yang et al., 2014). Briefly, protein samples were run on SDS–PAGE pre-cast gels (Bio-Rad), and were transferred to PVDF membranes. Membranes were blocked with TBST containing 5% milk, and incubated with a primary antibody overnight at 4 °C. After extensive washing, membranes were incubated with a secondary antibody for 1 h at room temperature. Proteins were then visualized using the enhanced chemiluminescence analysis system (GE Healthcare). After exposure, membranes were stripped and re-probed for β-actin as loading control. Image analysis was performed using the ImageJ program (NIH). The following primary antibodies were used: mouse monoclonal anti-SUMO1 (21C7; DSHB Hybridoma), rabbit polyclonal anti-SUMO2/3 (Covance), rabbit polyclonal anti-GFP (Invitrogen), and mouse monoclonal anti- β -actin (Sigma).
Immunofluorescence
Immunofluorescence staining was performed on paraffin-embedded brain sections (for SUMO1) or frozen brain sections (for SUMO2/3), as described previously (Yang et al., 2014). In short, after deparaffinization, brain sections (5-μm-thick) were incubated with primary antibodies at 4 °C overnight, followed by appropriate secondary fluorescent antibodies for 1 h at room temperature. The frozen brain sections (25 μm) were immunostained using a free-floating staining method. Confocal images were captured on a Leica SP5 confocal microscope (Leica Microsystems). The following primary antibodies were used: mouse monoclonal anti-SUMO1 (21C7; DSHB Hybridoma), rabbit polyclonal anti-SUMO2/3 (Covance), mouse monoclonal anti-GFP (Millipore), and rabbit polyclonal anti-GFP (Invitrogen).
Neurologic score system
Neurologic deficits were assessed by an observer blinded to the genotype of animals using a 9-point score system (Wellons et al., 2000). The assessment was performed before (Pre) and on day 4 after (Post) forebrain ischemia. After testing was complete, the score for each animal was the sum of the individual scores, with 0 = normal and 9 = severe injury.
Statistical analysis
The Prism 6 software (GraphPad) was used to analyze all data. Statistical analysis was assessed by Mann–Whitney U test on all data except the rCBF data. The rCBF data were analyzed by a 2-way analysis of variance (ANOVA) with Bonferroni's post hoc test for multiple comparisons. P values ≤0.05 were considered significant.
RESULTS
Effect of ischemia on SUMOylation in SUMO knockdown mice
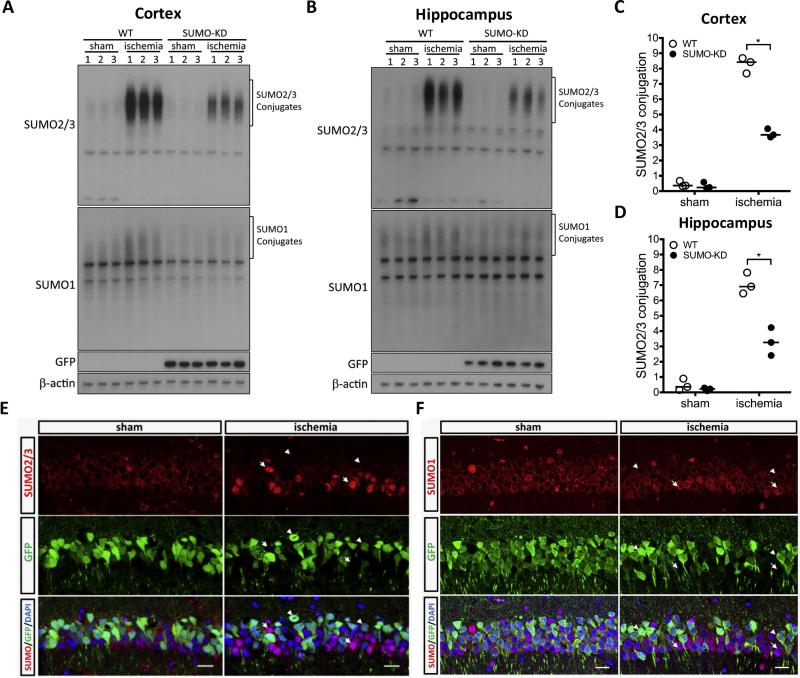
Detailed characterization of SUMO-KD mice has been described (Wang et al., 2014a). In this mouse model, three miRNAs that target SUMO1-3 are expressed together with GFP as indicator of transgene expression. Expression of designed miRNAs is driven by the Thy-1 promoter to achieve neuron-specific silencing of SUMO1-3. For the current study, we used SUMO-KD line 27 mice in which GFP is widely expressed in layer V of the cerebral cortex, and the hippocampal CA1 subfield (Wang et al., 2014a). Our earlier quantitative analysis revealed that in the hippocampal CA1 subfield, GFP is expressed in about 70% of neurons. To evaluate the change in SUMOylation after ischemia in SUMO-KD mice, we subjected wild-type and SUMO-KD mice to 10-min forebrain ischemia and 1 h reperfusion. Consistent with our previous findings (Yang et al., 2008c, 2014), levels of SUMO2/3-conjugated proteins in wild-type mice increased massively in both the cortex and hippocampus after brain ischemia (Fig. 1A, B). In SUMO-KD mice, however, the increase in post-ischemic SUMOylation was notably less (Fig. 1A, B). Quantitative Western blotting analysis indicated that the induction fold of SUMO2/3 conjugation in SUMO-KD was around 50% of that found in wild-type mice (Fig. 1C, D; Mann–Whitney U test, p ≤ 0.05, n = 3/group).
Fig. 1.
SUMOylation after transient forebrain ischemia in SUMO knockdown (SUMO-KD) mice. Wild-type (WT) and SUMO-KD mice were subjected to sham surgery or 10 10-min forebrain ischemia and 1 h reperfusion. Ischemia-induced changes in SUMOylation, and its subcellular localization were evaluated by Western blotting (A–D) and immunohistochemistry (E, F). (A–D) Global SUMOylation in cortex (A, C) and hippocampus (B, D). Global SUMOylation induced by ischemia/reperfusion was significantly less in SUMO-KD mice. The high-molecular-weight regions, marked by brackets, were used for quantification of SUMO2/3 conjugation. Intensities of SUMO2/3 conjugates were measured by image analysis, and normalized to β-actin. Horizontal bar = median values; *p ≤ 0.05. (E, F) Immunohistochemistry analysis of SUMO2/3 (E) and SUMO1 (F) in brains of SUMO-KD mice subjected to sham or ischemia surgery. After ischemia, strong nuclear SUMO2/3 staining was observed in GFP-negative hippocampus neurons (arrows), but was absent in SUMO knockdown neurons (GFP GFP-positive; arrowheads). Nuclear rim staining by SUMO1 appeared more intense in GFP GFP-negative neurons (arrows) compared to GFP GFP-positive cells (arrowheads). Scale bar = 20 μm.
We also used immunohistochemistry to evaluate the efficacy of SUMO1-3 miRNAs to silence SUMO expression (Fig. 1E, F). Double fluorescence immunostaining with anti-GFP and anti-SUMO2/3 antibodies revealed that the robust nuclear accumulation of SUMO2/3-conjugated proteins in post-ischemic GFP-negative neurons (Fig. 1E, arrows) was absent in GFP-positive cells (Fig. 1E, arrowheads). As expected, nuclear rim staining was observed for SUMO1. Unlike SUMO2/3, there was no dramatic increase in SUMO1-conjugated proteins nor obvious change in subcellular localization of SUMO1 after ischemia. However, the signal intensity of SUMO1 staining appeared less in GFP-positive neurons (Fig. 1F, arrowheads) than in GFP-negative neurons (Fig. 1F, arrows). Together, our data confirmed that the post-ischemic increase in SUMOylation was effectively suppressed in SUMO-KD mice.
Effects of ischemia on gene expression in SUMO knockdown mice
Our knowledge of the modulating effects of SUMO conjugation on gene expression is mostly based on results from in vitro cell culture studies. For this first genome-wide analysis of genes differentially regulated after brain ischemia in wild-type and SUMO-KD mice, we focused on the hippocampal CA1 subfield because CA1 neurons are particularly sensitive to transient ischemia, and SUMO expression is effectively silenced in most CA1 neurons of SUMO-KD mice (Fig. 1). Further, CA1 neurons play a key role in memory processes, and SUMO-KD mice are impaired in episodic and fear memory (Wang et al., 2014a). Since we have shown that the peak level of SUMO conjugated proteins in the brain induced by global forebrain ischemia is at 1 h of reperfusion (Yang et al., 2014), we decided to use brain samples collected at 3 h of reperfusion after ischemia for microarray analysis.
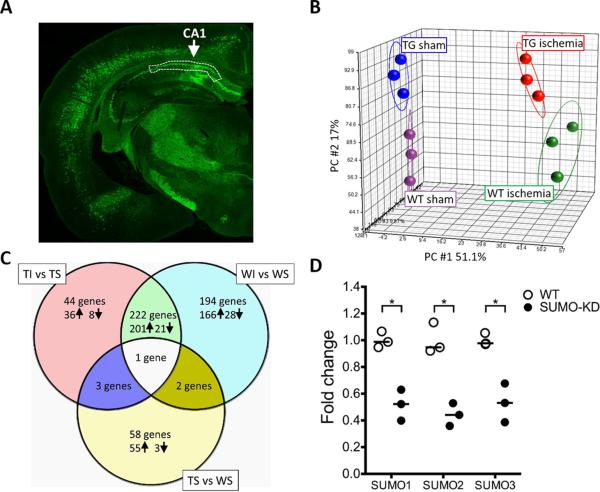
Microarray analysis was performed on samples prepared in triplicate from four groups: WS and WI, and SUMO-KD transgenic sham (TS) and ischemia (TI). The hippocampal CA1 subfield tissues were carefully excised, as indicated in Fig. 2A, and used for RNA preparation and microarray analysis. The raw data from the current study have been deposited in NCBI's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), and are accessible through GEO Series accession number GSE80681. An overview of microarray results is depicted in Fig. 2. First, we performed data analysis on the global gene expression profiles of all 12 samples by plotting individual samples in a 3-dimensional space based on principal components analysis (PCA; Fig. 2B). PCA demonstrated a clear separation between sham and ischemia, and between wild-type and SUMO-KD, indicating excellent reproducibility of samples and apparent effects of SUMO knockdown and ischemia on gene expression.
Fig. 2.
Overview of microarray data. Data analysis was performed on the global gene expression profiles of 12 samples from four groups: wild-type (WT) sham (WS), and ischemia (WI); and SUMO-KD (TG) sham (TS), and ischemia (TI). (A) The sampling regions. The region of hippocampal CA1 subfield that was dissected out and used for microarray analysis and qPCR is marked with white dot lines in a representative brain slice of SUMO-KD mice with GFP fluorescence. (B) Principal component analysis (PCA). The individual samples were plotted in a 3-dimensional space based on three principal components. Four groups of samples are clustered according to the genotype and surgery. (C) Venn diagram. The numbers of differentially regulated genes that were identified by pairwise comparisons of groups, with a cut-off of ≥ 2-twofold increase (↑) or decrease (↓) in gene expression are shown. (D) Verification of SUMO1-3 knockdown in SUMO-KD mice. The RNA samples from the sham group that were used for the microarray study, were analyzed to determine the levels of SUMO1-3 mRNA levels in WT and SUMO-KD mice. All individual data were normalized to β-actin. To calculate fold change, the mean values of WT mouse samples were set to 1.0. Horizontal bar = median values; *p ≤ 0.05.
We then defined selection criteria (≥ twofold change and p < 0.05) to identify genes differentially regulated in wild-type and SUMO-KD mice subjected to sham or ischemia surgery. The genes identified from different comparisons are listed in the Appendix Table and overviewed in Fig. 2C. To validate microarray data, we selected 11 of these genes, and performed qPCR analysis (Table 2). The same trend of expression change in the respective comparison, ie, up-regulation or down-regulation, was observed for all selected genes, although the fold changes of some genes were greater in the qPCR analysis than in the microarray data. Thus, the qPCR results validated our microarray data.
Table 2.
Verification of microarray data by qPCR analysis
| Entrez gene | Gene symbol | WI vs WS |
TI vs TS |
TS vs WS |
|||
|---|---|---|---|---|---|---|---|
| qPCR | Microarray | qPCR | Microarray | qPCR | Microarray | ||
| 11910 | Atf3 | 123.93 | 46.7 | 103.25 | 33.1 | −1.46 | −1.2 |
| 15507 | Hspb1 | 42.71 | 10.7 | 3.83 | 15.2 | 2.28 | −1.2 |
| 14281 | Fos | 39.58 | 11.4 | 64.74 | 20.2 | −1.79 | −1.8 |
| 11838 | Arc | 11.00 | 6.15 | 12.38 | 8.25 | −2.73 | −2.45 |
| 19735 | Rgs2 | 3.86 | 4.1 | 1.43 | 1.9 | −1.08 | −1.1 |
| 277353 | Tcfl5 | 3.36 | 1.56 | 0.83 | −1.27 | 261.38 | 38.58 |
| 20315 | Cxcl12 | −1.82 | −1.4 | −1.96 | −1.32 | −1.91 | −2.45 |
| 216233 | Socs2 | −1.36 | 1.95 | 1.17 | 2.18 | −2.40 | −1.1 |
| 108155 | Ogt | −2.21 | −2.3 | −1.89 | −2.2 | −0.96 | 1.2 |
| 28250 | Slco1a4 | −5.15 | −2.6 | −2.13 | −2.3 | −1.04 | −1.1 |
| 75668 | Rasl10a | −6.39 | −2.9 | −3.26 | −2.0 | −1.13 | −1.1 |
To characterize the effects of genotype and ischemia on gene regulation, we performed pairwise comparison of groups (Appendix Table). The number of differentially regulated genes among all 3 comparisons, and their expression changes after ischemia (up- or down-regulated) are presented in the Venn diagram in Fig. 2C A large number of genes were differentially regulated by ischemia in both wild-type and SUMO-KD mice. In wild-type mice, 419 genes were regulated by ischemia (Fig. 2C; WI vs WS), while 270 genes were regulated by ischemia in SUMO-KD mice (Fig. 2C; TI vs TS). However, a total of 196 genes showed ischemia-induced differential expression uniquely in wild-type mice, more than that in SUMO-KD mice. In sham animals, 64 genes were differently regulated by silencing SUMO expression, and most of these genes were up-regulated (Fig. 2C; TS vs WS). Notably, only one gene showed up in all three pairwise comparisons – the gene coding for the activity-regulated cytoskeleton-associated protein (Arc; also known as Arg3.1). Specifically, Arc was significantly down-regulated in SUMO-KD sham mice. After ischemia, it was up-regulated in both wild-type and SUMO-KD mice.
Microarray data show that the expression levels of SUMO1-3 decreased but with a fold change <2 in SUMO-KD mice compared to wild-type mice, and therefore, according to our pre-defined criteria, none of the three SUMOs were identified as differentially regulated genes (Appendix Table). To verify that expression of all SUMOs was indeed silenced in SUMO-KD mice, we performed qPCR on the hippocampal CA1 samples. The qPCR analysis revealed that SUMO1-3 mRNA levels in SUMO-KD mice were significantly reduced to 51.6% ± 6.7% (SUMO1), 44.3% ± 4.9% (SUMO2), and 53.1% ± 8.4% (SUMO3) compared to wild-type (Fig. 2D; mean ± SEM, Mann–Whitney U test, p ≤ 0.05, n = 3/group). This confirms the high capacity of SUMO miRNAs to silence SUMO expression, considering that SUMOs are expressed in all mammalian cells, and that here, SUMO is knocked down only in neurons of which about 70% express SUMO miRNAs in the hippocampal CA1 subfield. Taken together, the microarray analysis generated a reliable dataset of genes that are differentially regulated by SUMO conjugation in the CA1 subfield after brain ischemia.
Effects of ischemia on processes and pathways in SUMO knockdown mice
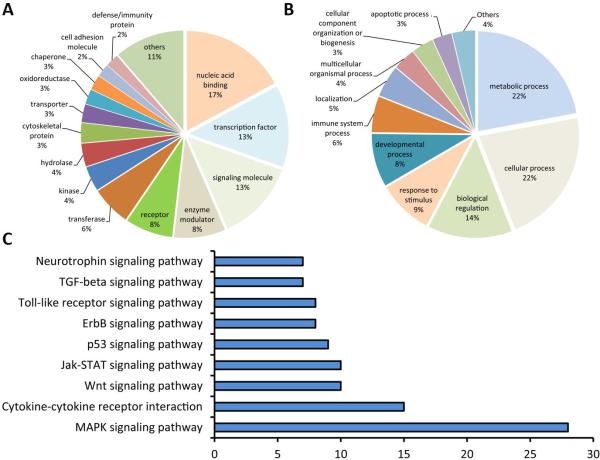
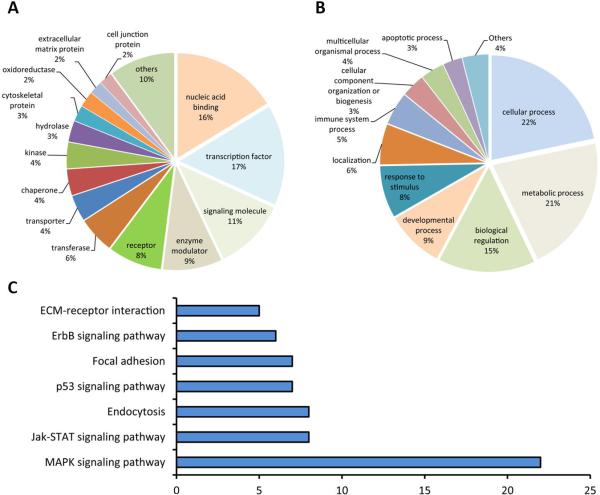
We then analyzed our microarray data based on two widely used databases – GO categories and KEGG pathways. GO and KEGG pathway analyses of differentially expressed genes in the CA1 subfield after forebrain ischemia in wild-type and SUMO-KD mice are depicted in Figs. 3 and 4, respectively. Overall, enriched GO terms according to protein class and biological processes, overlapped almost completely between wild-type and SUMO-KD mice. The list of enriched KEGG pathways show some overlaps but also some distinct differences between genotypes. In both datasets, the mitogen-activated protein kinase (MAPK) signaling pathway is particularly enriched, followed by Jak-STAT, p53, and ErbB signaling pathways (Figs. 3 and 4C and C). In these lists, ECM-receptor interaction, endocytosis, and focal adhesion pathways showed up only in SUMO-KD mice after ischemia (Fig. 4C), whereas other pathways, such as TGF-beta, Toll-like receptor, and Wnt signaling, were identified only in wild-type mice after ischemia (Fig. 3C).
Fig. 3.
GO classification and KEGG pathway analysis of differentially expressed genes in the CA1 samples after forebrain ischemia in wild-type mice. (A) Enriched GO terms according to protein class. (B) Enriched GO terms according to biological processes. (C) Enriched KEGG pathways.
Fig. 4.
GO classification and KEGG pathway analysis of differentially expressed genes in the CA1 samples after forebrain ischemia in SUMO-KD mice. (A) Enriched GO terms according to protein class. (B) Enriched GO terms according to biological processes. (C) Enriched KEGG pathways.
Gene expression in neurons before and after ischemia in SUMO knockdown mice
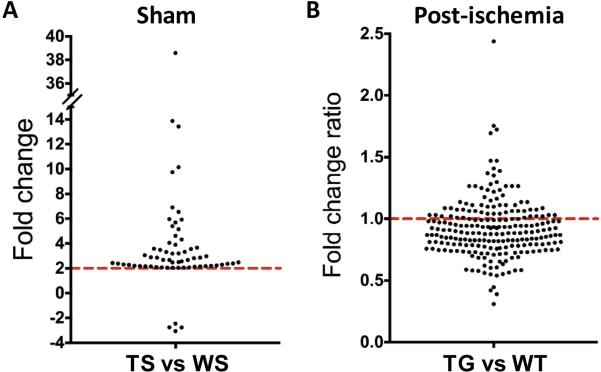
Our microarray results suggest that when SUMO expression is silenced in neurons, gene expression is differentially modulated in physiologic vs post-ischemic states (Fig. 2). Fig. 5 shows a visual representation of all genes that were differentially expressed in wild-type and SUMO-KD mice before (Fig. 5A) and after ischemia (Fig. 5B). Notably, of the 64 genes that were regulated by SUMOylation in the physiologic state, 60 were up-regulated, and only four were down-regulated in SUMO-KD mice (Fig. 5A). This observation supports a model whereby in non-stressed neurons SUMO conjugation negatively regulates gene expression in vivo. To better visualize the effect of SUMO conjugation on gene expression after ischemia, we compared the fold change of all 223 genes regulated by ischemia in both wild-type and SUMO-KD mice (Fig. 5B). Notably, about 70% of these genes showed a greater fold-change in wild-type compared to SUMO-KD mice (Fig. 5B, fold change ratio below 1), indicating that knockdown of SUMO expression suppresses the post-ischemic activation of gene expression.
Fig. 5.
Effect of SUMO knockdown on differentially expressed genes in the CA1 subfield before and after ischemia. (A) Fold change of genes differentially regulated in the SUMO-KD sham group (TS) compared to the wild-type sham group (WS). (B) The ratio of fold change of the genes differentially regulated by ischemia in both SUMO-KD (TG) and wild-type (WT) mice. The ratio was calculated by dividing an ischemia-induced fold change of a gene between TG and WT mice. If the ratio of a gene is < 1, the fold change of this gene regulated by ischemia is greater in WT mice.
Functional recovery after transient forebrain ischemia in SUMO knockdown mice
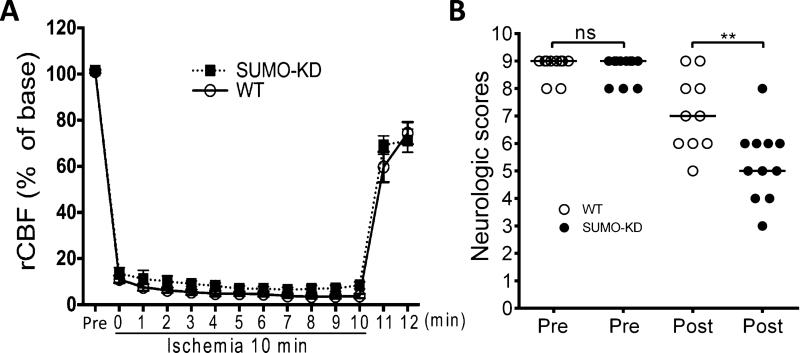
Results from a variety of in vitro and in vivo studies suggest that the post-ischemic activation of SUMO conjugation is a neuroprotective stress response. Recovery of neurologic function defines quality of life for patients who have suffered an ischemic attack; however, the effect of SUMO conjugation on functional recovery after ischemia has not yet been studied. To address this important aspect, we first confirmed that neuron-specific SUMO knockdown did not affect blood flow reduction in our transient forebrain ischemia model (Fig. 6A). Then, we used a 9-point scoring system to evaluate neurologic function before and 4 days after 10-min forebrain ischemia in wild-type and SUMO-KD mice. We did not find a significant difference in neurologic scores between wild-type and SUMO-KD mice before ischemia (Fig. 6B). After ischemia, however, impairment of neurologic functions was significantly more pronounced in SUMO-KD compared to wild-type mice (Fig. 6B; Mann–Whitney U test, p ≤ 0.01; n = 10 or 11/group).
Fig. 6.
Functional outcome after transient forebrain ischemia. (A) Effect of SUMO knockdown on rCBF in our transient forebrain ischemia model. Wild-type (WT; n = 5) and SUMO-KD (n = 5) mice were subjected to 10 10-min forebrain ischemia. Data are presented as means ± SEM. Laser-Doppler measurements revealed a similar change in rCBF between WT and SUMO-KD mice during ischemia and reperfusion. (B) Wild-type (WT; n = 10) and SUMO-KD (n = 11) mice were subjected to 10 10-min forebrain ischemia. Neurologic function was evaluated 1 day before (Pre) and 4 days after (Post) forebrain ischemia. Horizontal bar = median values. ns, not significant; **p ≤ 0.01.
DISCUSSION
Recently, SUMO conjugation has attracted attention in neuroscience, because this post-translational protein modification plays key roles in physiologic neurologic functions, including memory processes, and is associated with a variety of diseases of major clinical significance including brain tumor, brain ischemia, and neurodegenerative diseases (Yang et al., 2008b,c, 2013; Flotho and Melchior, 2013; Krumova and Weishaupt, 2013). Therefore, it is important to understand how SUMOylation impacts the physiologic/disease process under investigation. We know that many SUMO targets are nuclear proteins involved in gene expression, but there is little information about how SUMOylation modulates gene expression in vivo because prior studies have been conducted predominantly in vitro. The tools are now available to modulate SUMO in vivo only in cells that are targets of the physiologic/pathologic process under investigation. Recently, we developed a mouse model (SUMO-KD) in which expression of all three SUMO iso-forms is silenced specifically in neurons. This new mouse model has allowed us, for the first time, to study the effect of SUMO conjugation on gene expression in neurons in vivo, both in the physiologic state and in the diseases associated with SUMO conjugation, and to elucidate the role of SUMOylation in functional recovery of neurons exposed to ischemic stress.
Neurons are extremely sensitive to stress conditions; neurologic function is impaired in many diseases including brain ischemia/stroke, neurodegenerative diseases, and neuropsychiatric conditions. Further, post-ischemic recovery of neuronal function and the capacity of the brain to activate SUMO conjugation when challenged by ischemic stress decline with advanced age (Liu et al., 2016). Therefore, our new mouse model and the results presented here could be of broad interest. It is important to note that the gene expression profiles reported here were evaluated by microarray analysis on the hippocampal CA1 samples. Thus, these findings represent changes that occurred in all cell types present in the sample. However, the effects of SUMOylation on gene expression relate predominantly to neurons because we used a neuron-specific promoter to express SUMO1-3 miRNAs to silence SUMO expression.
For this in vivo study on the effects of SUMO conjugation on gene expression, we used brain ischemia as the pathologic state of interest because we have already shown that brain ischemia massively activates SUMO conjugation and nuclear accumulation of SUMO2/3-conjugated proteins (Yang et al., 2008b,c, 2014). Further, it is widely appreciated that the post-ischemic activation of SUMO conjugation is a protective stress response that shields neurons from injury, as evidenced in in vitro and in vivo studies (Lee et al., 2007, 2009, 2011, 2016; Datwyler et al., 2011). However, the effect of SUMO conjugation on post-ischemic neurologic function has not yet been investigated. The most important findings in the present study can be summarized as follows: (a) in the physiologic state of neurons, expression of most genes regulated by SUMOylation was activated by silencing SUMO expression; (b) in most cases, the effects of ischemia on gene expression were less pronounced in SUMO-KD than in wild-type mice; and (c) silencing SUMO expression worsened neurologic function after brain ischemia. These findings will be discussed in detail below.
Here, we reported a genomic study on hippocampal CA1 subfield tissue samples from mice, to identify genes regulated by ischemia. To date, only a few studies have applied the microarray technology to analyze gene expression modulated by global brain ischemia in samples excised from the CA1 subfield, whole hippocampus, or whole hemisphere (Jin et al., 2001; Kawahara et al., 2004; Yakubov et al., 2004; Feng et al., 2007; Buttner et al., 2009). For a comprehensive discussion see also Schmidt-Kastner, 2015 (Schmidt-Kastner, 2015). All of these studies used rat models, and only 2 studies used microarrays representing more than 10,000 genes (Feng et al., 2007; Buttner et al., 2009). Since these two studies used a post-ischemic time point for analysis similar to our study, we compared our list of genes regulated by ischemia with the lists from these studies. We found a large overlap of genes identified, including Atf3, Jun, Fos, Ptgs2, Gadd45, Hmox1, Hsp70, Hsp27, and also overlap of the highly enriched pathways, based on KEGG analysis, including MAPK, Wnt, TGF- β, and Toll-like receptor pathways. These comparisons support the validity of our dataset.
One of the key findings of our study reported here was that silencing SUMO expression in hippocampal CA1 neurons in vivo had opposite effects on global gene expression in the physiologic state vs the post-ischemia state. Silencing SUMO expression up-regulated gene expression in non-stressed neurons, but suppressed global gene expression responses induced by transient ischemia. At first glance, the observation that the global gene expression response induced by transient ischemia was suppressed in SUMO-KD mice was an unexpected finding, because until recently, it was generally believed that SUMOylation of most transcription factors negatively regulates their activity. Indeed, studies on a number of individual SUMO targets involved in transcription show that SUMOylation reduces the transcription activity of most of these targets (Chymkowitch et al., 2015). However, comprehensive characterization of transcriptional regulation by SUMOylation on chromatin using ChIP-seq techniques, revealed a more complex role for SUMOylation in gene expression.
Using human fibroblasts for ChIP-seq analysis, SUMOylated proteins were found at promoters of many genes involved in cell growth and proliferation, and inhibition of SUMOylation up-regulated transcription of those genes (Neyret-Kahn et al., 2013). Another study, however, reported that SUMOylation has a positive effect on transcription of genes in stressed cell (Seifert et al., 2015). Notably, using ChIP-seq and RNA-seq techniques, and heat shock as the stress condition, the authors found markedly increased binding of SUMO2-conjugated proteins to active DNA regulatory regions of many pro-survival genes. This observation supports the notion that stress-induced activation of SUMO2/3 conjugation is a protective stress response that shields stressed cells from damage by activating expression of pro-survival genes. Therefore, blocking this pro-survival response in SUMOKD mice may have contributed to the worse functional outcome after transient forebrain ischemia. Together, although our study reported here focused on terminally differentiated neurons in vivo, whereas earlier studies performed analyses on dividing cells in vitro, the modulating effects of SUMOylation on gene expression were similar, ie, suppressing gene expression in the physiologic state and activating expression of groups of genes when cells are stressed.
Considering the observation that in heat shock-stressed cells, SUMO2 conjugation activates expression of pro-survival genes, we analyzed our dataset to identify pro-survival genes with suppressed ischemia-induced activation in SUMO-KD mice. DAVID analysis identified eight anti-apoptosis genes of which seven were less activated in SUMO-KD mice after ischemia: Bag3, Cited2, Cflar, Bdnf, Hells, Myc, and Cebp β. For example, Bag3, which was identified as an ischemia-regulated gene in an earlier microarray study (Schmidt-Kastner et al., 2002), codes for a protein with anti-apoptosis function through interaction with Hsp70 (Rosati et al., 2011). Activation of the growth arrest and DNA damage 45 (Gadd45) gene family was also suppressed after ischemia in SUMO-KD mice. All 3 members of this gene family – Gadd45a, Gadd45b, and Gadd45g – have been identified as ischemia-regulated genes and they may play a protective role in brain ischemia injury (Chen et al., 1998; Sultan and Sweatt, 2013).
It is also noteworthy that Arc is one of the few genes that were down-regulated in SUMO-KD mice in the physiologic state. This could be a finding of significant interest, because Arc is critical for embryogenesis, and is a pivotal regulator of synaptic plasticity (Liu et al., 2000; Shepherd and Bear, 2011). Both processes are modulated by SUMOylation. The potential consequences of SUMO-modulated Arc expression in embryogenesis and synaptic plasticity should, therefore, be elucidated in future studies.
In conclusion, we used SUMO knockdown transgenic mice to perform the first in vivo analysis of global SUMOylation on the transcriptome in neurons regulated by transient forebrain ischemia. Notably, earlier studies that analyzed the effect of SUMOylation on the transcriptome were performed in vitro and exposed cells to a single stress such as heat shock. Further, we provide the first evidence that silencing SUMO expression worsened functional outcome after brain ischemia. Thus, the present findings, together with earlier reports on the role of SUMOylation in brain ischemia, support that SUMOylation is a neuroprotective response, at least in part, through transcriptional regulation of stress response genes.
Acknowledgments
We thank Pei Miao for her excellent technical support, and Kathy Gage for her excellent editorial contributions. This study was supported by American Heart Association grant 12SDG11950003 (to W.Y.), and by National Institutes of Health R01 grants NS081299 and NS097554 (to W.P.). X.L. is supported by the grant from the National Natural Science Foundation of China (#81471175).
Abbreviations
- ANOVA
analysis of variance
- Arc
activity-regulated cytoskeleton-associated protein
- GFP
green fluorescent protein
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
mitogen-activated protein kinase
- miRNA
microRNA
- OGD
oxygen/glucose deprivation
- PCA
principal component analysis
- qPCR
quantitative real-time PCR
- RanGAP1
Ran GTPase-activating protein-1
- rCBF
regional cerebral blood flow
- SUMO
small ubiquitin-like modifier
- SUMO-KD
SUMO1-3 knockdown
- WI
wild-type ischemia
- WS
wild-type sham
APPENDIX A
| WI vs WS (194 genes) | |||||||
|---|---|---|---|---|---|---|---|
| Probeset ID | Entrez Gene | Gene Symbol | Gene Title | RefSeq Transcript ID | WI vs WS | TI vs TS | TS vs WS |
| 1449827_at | 11595 | Acan | aggrecan | NM_007424 | 7.3 | 2.2 | −1.1 |
| 1422053_at | 16323 | Inhba | inhibin beta-A | NM_008380 | 5.4 | 1.2 | −1.4 |
| 1421009_at | 58185 | Rsad2 | radical S-adenosyl methionine domain containing 2 | NM_021384 | 5.3 | 2.6 | −1.0 |
| 1419220_at | 22437 | Xirp1 | xin actin-binding repeat containing 1 | NM_001081339 /// NM_011724 | 5.2 | 1.1 | −1.1 |
| 1437279_x_at | 20969 | Sdc1 | syndecan 1 | NM_011519 | 5.2 | 2.0 | 1.3 |
| 1418930_at | 15945 | Cxcl10 | chemokine (C-X-C motif) ligand 10 | NM_021274 | 5.1 | 2.3 | −1.1 |
| 1419247_at | 19735 | Rgs2 | regulator of G-protein signaling 2 | NM_009061 | 4.1 | 1.9 | −1.1 |
| 1423310_at | 21983 | Tpbg | trophoblast glycoprotein | NM_001164792 /// NM_011627 | 3.8 | 1.8 | 1.9 |
| 1424229_at | 226419 | Dyrk3 | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 | NM_145508 | 3.8 | 1.5 | 1.1 |
| 1418392_a_at | 55932 | Gbp3 | guanylate binding protein 3 | NM_018734 | 3.8 | 1.7 | 1.2 |
| 1435137_s_at | 319269 | 1200015M12Rik /// A130040M12Rik | RIKEN cDNA 1200015M12 gene /// RIKEN cDNA A130040M12 gene | NR_002860 | 3.7 | 2.8 | −1.2 |
| 1416579_a_at | 17075 | Epcam | epithelial cell adhesion molecule | NM_008532 | 3.6 | 1.8 | −1.0 |
| 1418203_at | 58801 | Pmaip1 | phorbol-12-myristate-13-acetate-induced protein 1 | NM_021451 | 3.6 | 1.8 | 1.7 |
| 1421134_at | 11839 | Areg | amphiregulin | NM_009704 | 3.5 | 1.4 | −1.0 |
| 1428781_at | 73712 | Dmkn | dermokine | NM_001166173 /// NM_001166174 /// NM_028618 /// NM_172899 | 3.5 | 1.3 | −1.2 |
| 1455271_at | 620695 | Gm13889 | predicted gene 13889 | NM_001145034 | 3.3 | 2.0 | 1.4 |
| 1418240_at | 14469 | Gbp2 | guanylate binding protein 2 | NM_010260 | 3.3 | 1.7 | −1.2 |
| 1417487_at | 14283 | Fosl1 | fos-like antigen 1 | NM_010235 | 3.2 | 1.9 | 1.1 |
| 1424711_at | 83921 | Tmem2 | transmembrane protein 2 | NM_001033759 /// NM_031997 | 3.2 | 1.6 | 1.2 |
| 1450698_at | 13537 | Dusp2 | dual specificity phosphatase 2 | NM_010090 | 3.2 | 1.7 | 1.0 |
| 1418538_at | 105785 | Kdelr3 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 | NM_134090 | 3.1 | 1.7 | −1.1 |
| 1420591_at | 80910 | Gpr84 | G protein-coupled receptor 84 | NM_030720 | 3.1 | 2.0 | −1.1 |
| 1419766_at | 17691 | Sik1 | salt inducible kinase 1 | NM_010831 | 3.1 | 1.9 | 1.2 |
| 1453238_s_at | 319269 | A130040M12Rik | RIKEN cDNA A130040M12 gene | NR_002860 | 3.0 | 2.5 | −1.2 |
| 1449824_at | 96875 | Prg4 | proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) | NM_001110146 /// NM_021400 | 3.0 | 2.1 | −1.5 |
| 1449484_at | 20856 | Stc2 | stanniocalcin 2 | NM_011491 | 2.9 | 2.3 | −1.3 |
| 1423233_at | 12609 | Cebpd | CCAAT/enhancer binding protein (C/EBP), delta | NM_007679 | 2.9 | 2.0 | 1.4 |
| 1424130_a_at | 19285 | Ptrf | polymerase I and transcript release factor | NM_008986 | 2.9 | 1.4 | −1.2 |
| 1427359_at | 338523 | Jhdm1d | jumonji C domain-containing histone demethylase 1 homolog D (S. cerevisiae) | NM_001033430 | 2.8 | 1.7 | −1.1 |
| 1449025_at | 15959 | Ifit3 | interferon-induced protein with tetratricopeptide repeats 3 | NM_010501 | 2.8 | 1.5 | 1.2 |
| 1449031_at | 12705 | Cited1 | Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 | NM_001276466 /// NM_001276473 /// NM_001276474 /// NM_007709 | 2.8 | 1.8 | 1.3 |
| 1450783_at | 15957 | Ifit1 | interferon-induced protein with tetratricopeptide repeats 1 | NM_008331 | 2.8 | 2.1 | −1.2 |
| 1417051_at | 18530 | Pcdh8 | protocadherin 8 | NM_001042726 /// NM_021543 | 2.8 | 1.5 | 1.0 |
| 1424594_at | 74480 | Samd4 | sterile alpha moif domain containing 4 | NM_001037221 /// NM_001163433 /// NM_028966 | 2.7 | 1.9 | −1.1 |
| 1424289_at | 209212 | Osgin2 | oxidative stress induced growth inhibitor family member 2 | NM_145950 | 2.7 | 1.8 | 1.1 |
| 1423703_at | 235036 | Ppan | peter pan homolog (Drosophila) | NM_145610 | 2.7 | 2.0 | 1.2 |
| 1423389_at | 17131 | Smad7 | SMAD family member 7 | NM_001042660 /// NM_008543 | 2.7 | 1.8 | −1.2 |
| 1430978_at | 75617 | Rps25 | ribosomal protein S25 | NM_024266 | 2.6 | 1.6 | 1.0 |
| 1428374_at | 93683 | Glce | glucuronyl C5-epimerase | NM_033320 | 2.6 | 1.6 | 1.3 |
| 1418135_at | 17355 | Aff1 | AF4/FMR2 family, member 1 | NM_001080798 /// NM_133919 | 2.6 | 1.5 | 1.6 |
| 1448961_at | 18828 | Plscr2 | phospholipid scramblase 2 | NM_001195084 /// NM_008880 | 2.6 | 2.0 | 1.2 |
| 1435655_at | 269261 | Rpl12 | ribosomal protein L12 | NM_009076 | 2.6 | 1.8 | 1.2 |
| 1440831_at | 12013 | Bach1 | BTB and CNC homology 1 | NM_007520 | 2.6 | 1.9 | 1.1 |
| 1452869_at | 66921 | Prpf38b | PRP38 pre-mRNA processing factor 38 (yeast) domain containing B | NM_025845 | 2.6 | 2.0 | −1.0 |
| 1448480_at | 66164 | Nip7 | nuclear import 7 homolog (S. cerevisiae) | NM_001164472 /// NM_025391 /// NR_028367 | 2.6 | 1.7 | 1.2 |
| 1418666_at | 19288 | Ptx3 | pentraxin related gene | NM_008987 | 2.6 | 1.9 | −1.2 |
| 1424932_at | 13649 | Egfr | epidermal growth factor receptor | NM_007912 /// NM_207655 | 2.6 | 1.9 | 1.3 |
| 1415996_at | 56338 | Txnip | thioredoxin interacting protein | NM_001009935 /// NM_023719 | 2.6 | 2.3 | 1.1 |
| 1421336_at | 19130 | Prox1 | prospero-related homeobox 1 | NM_008937 | 2.6 | 1.2 | 1.4 |
| 1416762_at | 20194 | S100a10 | S100 calcium binding protein A10 (calpactin) | NM_009112 | 2.5 | 1.9 | 1.2 |
| 1426648_at | 17164 | Mapkapk2 | MAP kinase-activated protein kinase 2 | NM_008551 | 2.5 | 1.6 | 1.0 |
| 1416123_at | 12444 | Ccnd2 | cyclin D2 | NM_009829 | 2.5 | 1.8 | 1.2 |
| 1429783_at | 56376 | Pdlim5 | PDZ and LIM domain 5 | NM_001190852 /// NM_001190853 /// NM_001190854 /// NM_001190855 /// NM_001190856 ///NM | 2.5 | 1.9 | 1.4 |
| 1426871_at | 70611 | Fbxo33 | F-box protein 33 | NM_001033156 | 2.5 | 1.9 | −1.1 |
| 1418492_at | 23893 | Grem2 | gremlin 2 homolog, cysteine knot superfamily (Xenopus laevis) | NM_011825 | 2.5 | 1.7 | 1.1 |
| 1450986_at | 55989 | Nop58 | NOP58 ribonucleoprotein | NM_018868 | 2.5 | 2.0 | 1.1 |
| 1425037_at | 224014 | Fgd4 | FYVE, RhoGEF and PH domain containing 4 | NM_139232 /// NM_139233 /// NM_139234 | 2.5 | 1.7 | 1.3 |
| 1418648_at | 112407 | Egln3 | EGL nine homolog 3 (C. elegans) | NM_028133 | 2.5 | 1.5 | −1.3 |
| 1424371_at | 228769 | Psmf1 | proteasome (prosome, macropain) inhibitor subunit 1 | NM_144889 /// NM_212446 | 2.5 | 1.7 | 1.4 |
| 1451714_a_at | 26397 | Map2k3 | mitogen-activated protein kinase kinase 3 | NM_008928 | 2.5 | 1.9 | 1.1 |
| 1460335_at | 80289 | Lysmd3 | LysM, putative peptidoglycan-binding, domain containing 3 | NM_030257 | 2.5 | 1.8 | 1.1 |
| 1424001_at | 67949 | Mki67ip | Mki67 (FHA domain) interacting nucleolar phosphoprotein | NM_026472 | 2.4 | 1.6 | 1.0 |
| 1420376_a_at | 15078 | Gm10257 /// Gm12657 /// Gm6749/// H3f3a /// H3f3b /// H3f3c /// LOC101056659 | predicted gene 10257 /// predicted gene 12657 /// predicted pseudogene 6749 /// H3 hist | NM_001081019 /// NM_008210 /// NM_008211 /// XM_003084942 /// XM_003945939 /// XM_00394 | 2.4 | 1.8 | 1.2 |
| 1421375_a_at | 20200 | S100a6 | S100 calcium binding protein A6 (calcyclin) | NM_011313 | 2.4 | 1.8 | 1.2 |
| 1452192_at | 234344 | Naf1 | nuclear assembly factor 1 homolog (S. cerevisiae) | NM_001163564 | 2.4 | 1.3 | 1.1 |
| 1434380_at | 229900 | Gbp7 | guanylate binding protein 7 | NM_001083312 /// NM_145545 | 2.4 | 1.5 | 1.2 |
| 1422706_at | 65112 | Pmepa1 | prostate transmembrane protein, androgen induced 1 | NM_022995 | 2.4 | 1.7 | 1.0 |
| 1425503_at | 14538 | Gcnt2 | glucosaminyl (N-acetyl) transferase 2, I-branching enzyme | NM_008105 /// NM_023887 /// NM_133219 | 2.4 | 1.8 | 1.4 |
| 1420380_at | 20296 | Ccl2 | chemokine (C-C motif) ligand 2 | NM_011333 | 2.4 | 1.8 | 1.1 |
| 1417639_at | 30805 | Slc22a4 | solute carrier family 22 (organic cation transporter), member 4 | NM_019687 | 2.4 | 1.6 | 1.3 |
| 1419169_at | 50772 | Mapk6 | mitogen-activated protein kinase 6 | NM_015806 /// NM_027418 | 2.4 | 1.9 | −1.1 |
| 1424355_a_at | 20467 | Sin3b | transcriptional regulator, SIN3B (yeast) | NM_001113248 /// NM_009188 | 2.4 | 1.8 | 1.1 |
| 1450718_at | 23921 | Sh2b2 | SH2B adaptor protein 2 | NM_018825 | 2.4 | 1.9 | 1.4 |
| 1460695_a_at | 72061 | 2010111I01Rik | RIKEN cDNA 2010111I01 gene | NM_028079 | 2.4 | 1.7 | 1.2 |
| 1434128_a_at | 232976 | Zfp574 | zinc finger protein 574 | NM_001168506 /// NM_175477 | 2.3 | 1.7 | 1.0 |
| 1417508_at | 30945 | Rnf19a | ring finger protein 19A | NM_013923 | 2.3 | 1.3 | 1.1 |
| 1437843_s_at | 71844 | Nupl1 | nucleoporin like 1 | NM_170591 | 2.3 | 1.5 | 1.5 |
| 1421365_at | 14313 | Fst | follistatin | NM_008046 | 2.3 | 1.6 | 1.1 |
| 1449317_at | 12633 | Cflar | CASP8 and FADD-like apoptosis regulator | NM_009805 /// NM_207653 | 2.3 | 1.7 | 1.1 |
| 1426351_at | 15510 | Hspd1 /// LOC101056370 | heat shock protein 1 (chaperonin) /// 60 kDa heat shock protein, mitochondrial-like | NM_010477 /// XM_003946023 | 2.3 | 1.9 | −1.0 |
| 1424107_at | 228421 | Kif18a | kinesin family member 18A | NM_139303 | 2.3 | 1.5 | 1.1 |
| 1418825_at | 15944 | Irgm1 | immunity-related GTPase family M member 1 | NM_008326 | 2.3 | 1.1 | 1.8 |
| 1451680_at | 76650 | Srxn1 | sulfiredoxin 1 homolog (S. cerevisiae) | NM_029688 | 2.3 | 1.4 | −1.0 |
| 1433502_s_at | 104662 | Tsr1 | TSR1 20S rRNA accumulation | NM_177325 | 2.3 | 1.7 | 1.2 |
| 1438761_a_at | 18263 | Odc1 | ornithine decarboxylase, structural 1 | NM_013614 | 2.3 | 1.5 | 1.2 |
| 1431422_a_at | 56405 | Dusp14 | dual specificity phosphatase 14 | NM_019819 | 2.3 | 1.5 | 1.3 |
| 1425671_at | 26556 | Homer1 | homer homolog 1 (Drosophila) | NM_011982 /// NM_147176 /// NM_152134 | 2.3 | 1.7 | −1.1 |
| 1452358_at | 24004 | Rai2 | retinoic acid induced 2 | NM_001103367 /// NM_198409 | 2.3 | 1.1 | 1.1 |
| 1422473_at | 18578 | Pde4b | phosphodiesterase 4B, cAMP specific | NM_001177980 /// NM_001177981 /// NM_001177982 /// NM_001177983 /// NM_019840 | 2.3 | 1.7 | 1.4 |
| 1426798_a_at | 108954 | Ppp1r15b | protein phosphatase 1, regulatory (inhibitor) subunit 15b | NM_133819 | 2.3 | 1.9 | 1.0 |
| 1418834_at | 117197 | Bloc1s4 | biogenesis of organelles complex-1, subunit 4, cappuccino | NM_133724 | 2.3 | 1.9 | 1.1 |
| 1449007_at | 12228 | Btg3 /// Gm7334 | B cell translocation gene 3 /// B-cell translocation gene 3 pseudogene | NM_009770 /// NR_002700 | 2.3 | 1.7 | 1.3 |
| 1419765_at | 71745 | Cul2 | cullin 2 | NM_029402 | 2.3 | 1.8 | 1.4 |
| 1425565_at | 19712 | Rest | RE1-silencing transcription factor | NM_011263 | |||
| 1424270_at | 13175 | Dclk1 | doublecortin-like kinase 1 | NM_001111051 /// NM_001111052 /// NM_001111053 /// NM_001195538 /// NM_001195539 /// NM | 2.2 | 1.4 | 1.0 |
| 1438992_x_at | 11911 | Atf4 | activating transcription factor 4 | NM_009716 | 2.2 | 1.9 | −1.2 |
| 1427321_s_at | 13052 | Cxadr | coxsackie virus and adenovirus receptor | NM_001025192 /// NM_001276263 /// NM_009988 | 2.2 | 1.7 | 1.3 |
| 1436684_a_at | 67045 | Riok2 | RIO kinase 2 (yeast) | NM_025934 | 2.2 | 1.5 | 1.5 |
| 1439154_at | 269966 | Nup98 | nucleoporin 98 | NM_022979 | 2.2 | 1.8 | 1.1 |
| 1435561_at | 13875 | Erf | Ets2 repressor factor | NM_010155 | 2.2 | 1.7 | 1.1 |
| 1449227_at | 12642 | Ch25h | cholesterol 25-hydroxylase | NM_009890 | 2.2 | 1.9 | 1.1 |
| 1448133_at | 97112 | Nmd3 | NMD3 homolog (S. cerevisiae) | NM_133787 | 2.2 | 1.6 | 1.2 |
| 1416106_at | 100087 | Kti12 | KTI12 homolog, chromatin associated (S. cerevisiae) | NM_029571 | 2.2 | 1.6 | 1.1 |
| 1415806_at | 18791 | Plat | plasminogen activator, tissue | NM_008872 | 2.2 | 1.8 | 1.2 |
| 1417719_at | 60406 | Sap30 | sin3 associated polypeptide | NM_021788 | 2.2 | 1.9 | 1.1 |
| 1421267_a_at | 17684 | Cited2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | NM_010828 | 2.2 | 1.4 | 1.6 |
| 1450297_at | 16193 | Il6 | interleukin 6 | NM_031168 | 2.2 | 1.8 | 1.0 |
| 1434436_at | 75746 | Morc4 | microrchidia 4 | NM_001193309 /// NM_029413 | 2.2 | 1.8 | 1.1 |
| 1419702_at | 21339 | Taf1a | TATA box binding protein (Tbp)-associated factor, RNA polymerase I, A | NM_001277957 /// NM_001277958 /// NM_001277959 /// NM_021466 | 2.2 | 1.5 | −1.1 |
| 1421855_at | 14190 | Fgl2 | fibrinogen-like protein 2 | NM_008013 | 2.2 | 1.3 | 1.2 |
| 1452997_at | 381598 | 2610005L07Rik | cadherin 11 pseudogene | NM_001024708 /// NM_001033456 /// NR_028428 | 2.2 | 1.5 | 1.5 |
| 1460351_at | 20195 | Gm12854 /// Gm5068 /// S100a11 | predicted gene 12854 /// predicted gene 5068 /// S100 calcium binding protein A11 (calg | NM_016740 /// XM_003085822 /// XM_003086388 /// XM_204772 | 2.2 | 1.8 | −1.1 |
| 1448328_at | 24055 | Sh3bp2 | SH3-domain binding protein 2 | NM_001136088 /// NM_001145858 /// NM_001145859 /// NM_011893 | 2.2 | 2.0 | 1.3 |
| 1420502_at | 20229 | Sat1 | spermidine/spermine N1-acetyl transferase 1 | NM_009121 | 2.2 | 1.9 | −1.0 |
| 1459902_at | 212772 | Arl14ep | ADP-ribosylation factor-like 14 effector protein | NM_001025102 /// NM_173750 | 2.2 | 1.8 | 1.0 |
| 1451533_at | 217887 | BC022687 | cDNA sequence BC022687 | NM_145450 | 2.2 | 1.5 | 1.5 |
| 1451969_s_at | 235587 | Parp3 | poly (ADP-ribose) polymerase family, member 3 | NM_145619 | 2.2 | 2.0 | −1.1 |
| 1418804_at | 84112 | Sucnr1 | succinate receptor 1 | NM_032400 | 2.2 | 1.2 | 1.0 |
| 1448678_at | 73225 | Fam118a | family with sequence similarity 118, member A | NM_133750 /// NM_177067 | 2.2 | 1.9 | 1.1 |
| 1423169_at | 24074 | Taf7 | TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor | NM_175770 | 2.1 | 1.6 | 1.6 |
| 1455105_at | 19248 | Ptpn12 | protein tyrosine phosphatase, non-receptor type 12 | NM_011203 | 2.1 | 1.5 | −1.0 |
| 1452416_at | 16194 | Il6ra | interleukin 6 receptor, alpha | NM_010559 | 2.1 | 1.9 | 1.2 |
| 1424211_at | 70556 | Slc25a33 | solute carrier family 25, member 33 | NM_027460 /// XM_003945705 | 2.1 | 1.4 | 1.3 |
| 1451780_at | 17060 | Blnk | B cell linker | NM_008528 | 2.1 | 1.6 | −1.1 |
| 1460428_at | 68420 | Ankrd13a | ankyrin repeat domain 13a | NM_026718 | 2.1 | 1.8 | 1.0 |
| 1452045_at | 226442 | Zfp281 | zinc finger protein 281 | NM_001160251 /// NM_177643 | 2.1 | 1.9 | −1.0 |
| 1436893_a_at | 57438 | 41705 | membrane-associated ring finger (C3HC4) 7 | NM_020575 | 2.1 | 1.6 | 1.3 |
| 1422554_at | 66647 | Ndnl2 | necdin-like 2 | NM_023239 | 2.1 | 1.8 | −1.0 |
| 1460657_at | 22409 | Wnt10a | wingless related MMTV integration site 10a | NM_009518 | 2.1 | 1.3 | 1.1 |
| 1418711_at | 18590 | Pdgfa | platelet derived growth factor, alpha | NM_008808 | 2.1 | 1.7 | −1.0 |
| 1454064_a_at | 56515 | Rnf138 | ring finger protein 138 | NM_019706 /// NM_207623 | 2.1 | 1.5 | 1.2 |
| 1448749_at | 56193 | Plek | pleckstrin | NM_019549 | 2.1 | 1.9 | 1.1 |
| 1451069_at | 223775 | Pim3 | proviral integration site 3 | NM_145478 | 2.1 | 1.9 | −1.1 |
| 1426858_at | 16324 | Inhbb | inhibin beta-B | NM_008381 | 2.1 | 1.6 | 1.6 |
| 1416359_at | 170625 | Snx18 | sorting nexin 18 | NM_130796 | 2.1 | 2.0 | −1.1 |
| 1437111_at | 244871 | Zc3h12c | zinc finger CCCH type containing 12C | NM_001162921 | 2.1 | 1.8 | −1.1 |
| 1426609_at | 72662 | Dis3 | DIS3 mitotic control homolog (S. cerevisiae) | NM_028315 | 2.1 | 1.8 | 1.1 |
| 1450685_at | 59046 | Arpp19 | cAMP-regulated phosphoprotein 19 | NM_001142655 /// NM_021548 | 2.1 | 1.5 | −1.2 |
| 1416081_at | 17125 | Smad1 | SMAD family member 1 | NM_008539 | 2.1 | 1.9 | 1.1 |
| 1425321_a_at | 94040 | Clmn | calmin | NM_001040682 /// NM_053155 | 2.1 | 1.9 | 1.1 |
| 1418918_at | 16006 | Igfbp1 | insulin-like growth factor binding protein 1 | NM_008341 | 2.1 | 1.5 | 1.1 |
| 1423904_a_at | 52118 | Pvr | poliovirus receptor | NM_027514 | 2.1 | 1.5 | 1.2 |
| 1448802_at | 27275 | Nufip1 | nuclear fragile X mental retardation protein interacting protein 1 | NM_013745 | 2.1 | 1.7 | 1.1 |
| 1434425_at | 99681 | Tchh | trichohyalin | NM_001163098 | 2.1 | 1.3 | 1.4 |
| 1428114_at | 108052 | Slc14a1 | solute carrier family 14 (urea transporter), member 1 | NM_001171010 /// NM_001171011 /// NM_028122 | 2.1 | 1.3 | 1.7 |
| 1437696_at | 381066 | Zfp948 | zinc finger protein 948 | NM_001002008 | 2.1 | 1.4 | 1.1 |
| 1428942_at | 17750 | Mt2 | metallothionein 2 | NM_008630 | 2.1 | 1.8 | 1.1 |
| 1429040_at | 547150 | 6820431F20Rik | cadherin 11 pseudogene | NR_030708 | 2.1 | 1.3 | 1.8 |
| 1418323_at | 14155 | Fem1b | feminization 1 homolog b (C. elegans) | NM_010193 | 2.1 | 1.9 | 1.0 |
| 1438511_a_at | 66214 | Rgcc | regulator of cell cycle | NM_025427 | 2.0 | 1.3 | 1.1 |
| 1449188_at | 59090 | Midn | midnolin | NM_021565 | 2.0 | 1.7 | −1.1 |
| 1453100_at | 13000 | Csnk2a2 | casein kinase 2, alpha prime polypeptide | NM_009974 | 2.0 | 1.7 | 1.1 |
| 1419749_at | 13434 | Trdmt1 | tRNA aspartic acid methyltransferase 1 | NM_010067 | 2.0 | 1.7 | 1.3 |
| 1436871_at | 225027 | Srsf7 | serine/arginine rich splicing factor 7 - |
NM_001195485 /// NM_001195486 /// NM_001195487 /// NM_146083 /// NR_036615 | 2.0 | 1.7 | 1.3 |
| 1437396_at | 208647 | Creb3l2 | cAMP responsive element binding protein 3-like 2 | NM_178661 | 2.0 | 1.7 | 1.0 |
| 1460179_at | 15502 | Dnaja1 | DnaJ (Hsp40) homolog, subfamily A, member 1 | NM_001164671 /// NM_001164672 /// NM_008298 | 2.0 | 1.8 | −1.1 |
| 1416751_a_at | 53975 | Ddx20 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 | NM_017397 | 2.0 | 1.4 | 1.1 |
| 1415940_at | 100494 | Zfand2a | zinc finger, AN1-type domain 2A | NM_001159908 /// NM_133349 | 2.0 | 1.9 | 1.1 |
| 1420930_s_at | 54366 | Ctnnal1 | catenin (cadherin associated protein), alpha-like 1 | NM_018761 | 2.0 | 1.7 | 1.1 |
| 1431057_a_at | 76453 | Prss23 | protease, serine, 23 | NM_029614 | 2.0 | 1.3 | 1.1 |
| 1417426_at | 19073 | Srgn | serglycin | NM_011157 | 2.0 | 1.7 | −1.1 |
| 1449858_at | 12524 | Cd86 | CD86 antigen | NM_019388 | 2.0 | 1.7 | −1.0 |
| 1448413_at | 71952 | 2410016O06Rik | RIKEN cDNA 2410016O06 gene | NM_023633 | 2.0 | 1.5 | 1.0 |
| 1420664_s_at | 19124 | Procr | protein C receptor, endothelial | NM_011171 | 2.0 | 1.7 | −1.0 |
| 1419940_at | 109260 | C030018P15Rik | RIKEN cDNA C030018P15 gene | --- | 2.0 | 1.9 | −1.1 |
| 1437630_at | 224092 | Lsg1 | large subunit GTPase 1 homolog (S. cerevisiae) | NM_178069 | 2.0 | 1.4 | 1.2 |
| 1437320_s_at | 22590 | Xpa | xeroderma pigmentosum, complementation group A | NM_011728 | −2.0 | −1.9 | −1.0 |
| 1439453_x_at | 68209 | Rnaseh2c | ribonuclease H2, subunit C | NM_026616 | −2.0 | −1.8 | −1.1 |
| 1416053_at | 16979 | Lrrn1 | leucine rich repeat protein 1, neuronal | NM_008516 | −2.0 | −1.9 | 1.0 |
| 1429177_x_at | 20671 | Sox17 | SRY-box containing gene 17 | NM_011441 | −2.0 | −1.9 | 1.1 |
| 1419332_at | 54156 | Egfl6 | EGF-like-domain, multiple 6 | NM_019397 | −2.0 | −1.4 | 1.0 |
| 1426544_a_at | 67120 | Ttc14 | tetratricopeptide repeat domain 14 | NM_025978 /// NM_027619 | −2.0 | −1.8 | 1.2 |
| 1450803_at | 18205 | Ntf3 | neurotrophin 3 | NM_001164034 /// NM_001164035 /// NM_008742 | −2.1 | −1.6 | −1.3 |
| 1428483_a_at | 66578 | Mis18a | MIS18 kinetochore protein homolog A (S. pombe) | NM_025642 | −2.1 | −1.6 | −1.0 |
| 1455092_at | 22680 | Zfp207 | zinc finger protein 207 | NM_001130169 /// NM_001130170 /// NM_001130171 /// NM_011751 /// NR_045038 | −2.1 | −1.8 | 1.1 |
| 1448530_at | 66355 | Gmpr | guanosine monophosphate reductase | NM_025508 | −2.1 | −1.4 | −1.2 |
| 1455831_at | 233908 | Fus | fused in sarcoma | NM_139149 | −2.1 | −1.7 | −1.1 |
| 1425111_at | 66673 | Sorcs3 | sortilin-related VPS10 domain containing receptor 3 | NM_025696 | −2.1 | −1.3 | −1.4 |
| 1451901_at | 20585 | Hltf | helicase-like transcription factor | NM_009210 /// NM_144959 | −2.1 | −1.7 | 1.0 |
| 1418266_at | 11686 | Alox12b | arachidonate 12-lipoxygenase, 12R type | NM_009659 | −2.1 | −1.4 | 1.5 |
| 1423084_at | 26878 | B3galt2 | UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 | NM_020025 | −2.1 | −1.7 | 1.1 |
| 1438710_at | 15550 | Htr1a | 5-hydroxytryptamine (serotonin) receptor 1A | NM_008308 | −2.1 | −1.5 | −1.2 |
| 1425175_at | 227580 | C1ql3 | C1q-like 3 | NM_153155 | −2.1 | −1.7 | 1.0 |
| 1421818_at | 12053 | Bcl6 | B cell leukemia/lymphoma 6 | NM_009744 | −2.1 | −1.7 | −1.3 |
| 1438034_at | 66302 | Rmdn1 | regulator of microtubule dynamics 1 | NM_025476 | −2.2 | −1.9 | 1.1 |
| 1451344_at | 231633 | Tmem119 | transmembrane protein 119 | NM_146162 | −2.2 | −1.9 | −1.0 |
| 1425344_at | 67608 | Narf | nuclear prelamin A recognition factor | NM_026272 | −2.2 | −1.6 | −1.1 |
| 1417574_at | 20315 | Cxcl12 | chemokine (C-X-C motif) ligand 12 | NM_001012477 /// NM_013655 /// NM_021704 | −2.2 | −1.8 | −1.2 |
| 1428052_a_at | 68310 | Zmym1 | zinc finger, MYM domain containing 1 | NM_026670 | −2.2 | −1.9 | 1.1 |
| 1427017_at | 212712 | Satb2 | special AT-rich sequence binding protein 2 | NM_139146 | −2.3 | 1.2 | −1.4 |
| 1438465_at | 414758 | 5830428H23Rik | RIKEN cDNA 5830428H23 gene | NM_001001737 /// XR_105997 | −2.4 | −1.8 | −1.2 |
| 1426552_a_at | 14025 | Bcl11a | B cell CLL/lymphoma 11A (zinc finger protein) | NM_001159289 /// NM_001159290 /// NM_001242934 /// NM_016707 | −2.4 | −1.7 | −1.2 |
| 1424303_at | 211896 | Depdc7 | DEP domain containing 7 | NM_144804 | −2.5 | −1.2 | −2.0 |
| 1427975_at | 75668 | Rasl10a | RAS-like, family 10, member A | NM_145216 | −2.9 | −2.0 | −1.1 |
| TI vs TS (44 genes) | |||||||
|---|---|---|---|---|---|---|---|
| Probeset ID | Entrez Gene | Gene Symbol | Gene Title | ReSeq Transcript ID | WI vs WS | TI vs TS | TS vs WS |
| 1454770_at | 12426 | Cckbr | cholecystokinin B receptor | NM_007627 | 1.48 | 2.71 | −1.54 |
| 1450700_at | 260409 | Cdc42ep3 | CDC42 effector protein (Rho GTPase binding) 3 | NM_026514 | 1.58 | 2.48 | 1.10 |
| 1450843_a_at | 12406 | Serpinh1 | serine (or cysteine) peptidase inhibitor, clade H, member 1 | NM_001111043 /// NM_001111044 /// NM_009825 | 1.85 | 2.43 | −1.38 |
| 1423614_at | 100604 | Lrrc8c | leucine rich repeat containing 8 family, member C | NM_133897 | 1.68 | 2.43 | −1.25 |
| 1416897_at | 80285 | Parp9 | poly (ADP-ribose) polymerase family, member 9 | NM_030253 | 1.89 | 2.40 | −1.03 |
| 1417812_a_at | 16780 | Lamb3 | laminin, beta 3 | NM_001277928 /// NM_008484 | 1.96 | 2.40 | 1.17 |
| 1419654_at | 21887 | Tle3 | transducin-like enhancer of split 3, homolog of Drosophila E(spl) | NM_001083927 /// NM_001083928 /// NM_009389 | 1.94 | 2.37 | −1.07 |
| 1417332_at | 19725 | Rfx2 | regulatory factor X, 2 (influences HLA class II expression) | NM_009056 /// NM_027787 | 1.67 | 2.37 | −1.25 |
| 1420589_at | 15118 | Has3 | hyaluronan synthase 3 | NM_008217 | 1.44 | 2.33 | −1.80 |
| 1417240_at | 22793 | Zyx | zyxin | NM_011777 | 1.96 | 2.29 | −1.05 |
| 1449161_at | 13615 | Edn2 | endothelin 2 | NM_007902 | 1.79 | 2.29 | −1.27 |
| 1448228_at | 16948 | Lox | lysyl oxidase | NM_010728 | 1.10 | 2.27 | 1.59 |
| 1451794_at | 319880 | Tmcc3 | transmembrane and coiled coil domains 3 | NM_001168684 /// NM_172051 /// NM_177026 | 2.00 | 2.27 | −1.04 |
| 1451751_at | 73284 | Ddit4l | DNA-damage-inducible transcript 4-like | NM_030143 | 1.59 | 2.26 | −1.34 |
| 1418025_at | 20893 | Bhlhe40 | basic helix-loop-helix family, member e40 | NM_011498 | 1.86 | 2.24 | −1.25 |
| 1416287_at | 19736 | Rgs4 | regulator of G-protein signaling 4 | NM_009062 | 1.92 | 2.23 | −1.30 |
| 1415874_at | 24063 | Spry1 | sprouty homolog 1 (Drosophila) | NM_011896 | 1.42 | 2.21 | −1.25 |
| 1451873_a_at | 17129 | Smad5 | SMAD family member 5 | NM_001164041 /// NM_001164042 /// NM_008541 | 1.73 | 2.19 | −1.09 |
| 1425420_s_at | 50523 | Lats2 | large tumor suppressor 2 | NM_015771 /// NM_153382 | 1.97 | 2.17 | 1.04 |
| 1450350_a_at | 81703 | Jdp2 | Jun dimerization protein 2 | NM_001205052 /// NM_001205053 /// NM_030887 | 1.63 | 2.14 | −1.58 |
| 1431734_a_at | 67035 | Dnajb4 | DnaJ (Hsp40) homolog, subfamily B, member 4 | NM_025926 /// NM_027287 | 1.95 | 2.14 | −1.01 |
| 1448860_at | 140743 | Rem2 | rad and gem related GTP binding protein 2 | NM_080726 | 1.46 | 2.13 | −1.57 |
| 1450229_at | 26896 | Med14 | mediator complex subunit 14 | NM_001048208 /// NM_012005 | 1.83 | 2.12 | −1.03 |
| 1417884_at | 104681 | Slc16a6 | solute carrier family 16 (monocarboxylic acid transporters), member 6 | NM_001029842 /// NM_134038 | 1.35 | 2.12 | −1.18 |
| 1453678_at | 17190 | Mbd1 | methyl-CpG binding domain protein 1 | NM_013594 | 1.82 | 2.11 | 1.02 |
| 1417379_at | 29875 | Iqgap1 | IQ motif containing GTPase activating protein 1 | NM_016721 | 1.90 | 2.09 | −1.02 |
| 1416564_at | 20680 | Sox7 | SRY-box containing gene 7 | NM_011446 | 1.51 | 2.09 | −1.30 |
| 1416360_at | 170625 | Snx18 | sorting nexin 18 | NM_130796 | 1.98 | 2.08 | −1.12 |
| 1434901_at | 381990 | Zbtb2 | zinc finger and BTB domain containing 2 | NM_001033466 | 1.75 | 2.08 | −1.33 |
| 1449545_at | 14172 | Fgf18 | fibroblast growth factor 18 | NM_008005 /// NR_102395 | 1.77 | 2.08 | 1.14 |
| 1453392_at | 69863 | Ttc39b | tetratricopeptide repeat domain 39B | NM_025782 /// NM_027238 | 1.58 | 2.05 | −1.22 |
| 1450734_at | 89867 | Sec16b | SEC16 homolog B (S. cerevisiae) | NM_001159986 /// NM_033354 /// NR_027641 | 1.33 | 2.02 | −1.20 |
| 1451021_a_at | 12224 | Klf5 | Kruppel-like factor 5 | NM_009769 | 1.48 | 2.02 | −1.14 |
| 1418255_s_at | 20807 | Srf | serum response factor | NM_020493 | 1.94 | 2.01 | −1.38 |
| 1425400_a_at | 56222 | Cited4 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 | NM_019563 | 1.87 | 2.01 | 1.73 |
| 1439079_a_at | 59079 | Erbb2ip | Erbb2 interacting protein | NM_001005868 /// NM_021563 | 1.58 | 2.00 | −1.03 |
| 1449482_at | 382522 | Hist3h2ba /// Hist3h2bb-ps | histone cluster 3, H2ba /// histone cluster 3, H2bb, pseudogene | NM_030082 /// NM_206882 | 0.51 | −2.03 | 1.29 |
| 1416174_at | 26450 | Rbbp9 | retinoblastoma binding protein 9 | NM_015754 | 0.54 | −2.04 | 1.17 |
| 1451832_at | 75458 | Cklf | chemokine-like factor | NM_001037840 /// NM_001037841 /// NM_029295 /// NM_029313 | 0.60 | −2.18 | 1.54 |
| 1418788_at | 21687 | Tek | endothelial-specific receptor tyrosine kinase | NM_013690 | 0.52 | −2.26 | 1.01 |
| 1454858_x_at | 70152 | Mettl7a1 | methyltransferase like 7A1 | NM_027334 | 0.62 | −2.42 | 1.64 |
| 1437461_s_at | 67225 | Rnpc3 | RNA-binding region (RNP1, RRM) containing 3 | NM_001038696 /// NM_026043 | 0.50 | −2.47 | 1.10 |
| 1420048_at | 97455 | C78859 | expressed sequence C78859 | --- | 0.61 | −2.56 | 1.63 |
| 1456010_x_at | 15208 | Hes5 | hairy and enhancer of split 5 (Drosophila) | NM_010419 | 0.54 | −2.63 | 1.74 |
| TS vs WS (58 genes) | |||||||
|---|---|---|---|---|---|---|---|
| Probeset ID | Entrez Gene | Gene Symbol | Gene Title | RefSeq Transcript ID | WI vs WS | TI vs TS | TS vs WS |
| 1456515_s_at | 277353 | Tcfl5 | transcription factor-like 5 (basic helix-loop-helix) | NM_178254 | 1.59 | −1.28 | 38.59 |
| 1433579_at | 238257 | Tmem30b | transmembrane protein 30B | NM_178715 | 1.03 | −1.11 | 13.88 |
| 1417797_a_at | 69073 | 1810019J16Rik | RIKEN cDNA 1810019J16 gene | NM_001083916 /// NM_133707 | −1.02 | −1.55 | 13.43 |
| 1422825_at | 27220 | Cartpt | CART prepropeptide | NM_001081493 /// NM_013732 | −1.13 | −1.15 | 10.15 |
| 1424525_at | 225642 | Grp | gastrin releasing peptide | NM_175012 | 1.27 | −1.21 | 9.75 |
| 1416121_at | 16948 | Lox | lysyl oxidase | NM_010728 | 1.65 | 1.25 | 6.91 |
| 1418304_at | 170677 | Cdhr1 | cadherin-related family member 1 | NM_130878 | 1.28 | −1.48 | 6.55 |
| 1427509_at | 545192 | Baiap3 | BAI1-associated protein 3 | NM_001163270 | 1.45 | −1.53 | 5.96 |
| 1418941_at | 93893 | Pcdhb22 | protocadherin beta 22 | NM_053147 | 1.09 | −1.63 | 5.93 |
| 1420422_at | 93892 | Pcdhb21 | protocadherin beta 21 | NM_053146 | −1.28 | −1.81 | 5.15 |
| 1417680_at | 16493 | Kcna5 | potassium voltage-gated channel, shaker-related subfamily, member 5 | NM_145983 | 1.33 | −1.09 | 4.62 |
| 1417430_at | 12585 | Cdr2 | cerebellar degeneration-related 2 | NM_007672 | 1.56 | −1.04 | 4.32 |
| 1422530_at | 19132 | Prph | peripherin | NM_001163588 /// NM_001163589 /// NM_013639 | 1.07 | −1.36 | 4.05 |
| 1417920_at | 93835 | Amn | amnionless | NM_033603 | 1.74 | 1.32 | 3.90 |
| 1416627_at | 20732 | Spint1 | serine protease inhibitor, Kunitz type 1 | NM_016907 | 1.34 | −1.53 | 3.58 |
| 1421396_at | 18548 | Pcsk1 | proprotein convertase subtlisin/kexin type 1 | NM_013628 | 1.06 | −1.60 | 3.55 |
| 1418301_at | 54139 | Irf6 | interferon regulatory factor 6 | NM_016851 /// NM_178083 | −1.65 | −1.13 | 3.48 |
| 1449283_a_at | 29857 | Mapk12 | mitogen-activated protein kinase 12 | NM_013871 | 1.04 | −1.18 | 3.33 |
| 1452270_s_at | 65969 | Cubn | cubilin (intrinsic factor-cobalamin receptor) | NM_001081084 | 1.92 | −1.32 | 3.20 |
| 1449491_at | 105844 | Card10 | caspase recruitment domain family, member 10 | NM_130859 | 1.05 | −1.27 | 3.19 |
| 1422586_at | 13599 | Ecel1 | endothelin converting enzyme-like 1 | NM_001277925 /// NM_021306 | −1.16 | −1.25 | 3.05 |
| 1421129_a_at | 53313 | Atp2a3 | ATPase, Ca++ transporting, ubiquitous | NM_001163336 /// NM_001163337 /// NM_016745 | −1.01 | −1.64 | 2.97 |
| 1449583_at | 93891 | Pcdhb20 | protocadherin beta 20 | NM_053145 | −1.57 | −1.96 | 2.89 |
| 1425288_at | 231004 | Samd11 | sterile alpha motif domain containing 11 | NM_001110516 /// NM_173736 | 1.29 | 1.10 | 2.86 |
| 1419021_at | 109904 | Mcf2 | mcf.2 transforming sequence | NM_133197 | −1.38 | −1.33 | 2.86 |
| 1418488_s_at | 72388 | Ripk4 | receptor-interacting serine-threonine kinase 4 | NM_023663 | 1.25 | 1.41 | 2.82 |
| 1434674_at | 17101 | Lyst | lysosomal trafficking regulator | NM_010748 | −1.05 | −1.35 | 2.68 |
| 1418373_at | 56012 | Pgam2 | phosphoglycerate mutase 2 | NM_018870 | −1.39 | −1.71 | 2.67 |
| 1426271_at | 226026 | Smc5 | structural maintenance of chromosomes 5 | NM_001252684 /// NM_001252685 /// NM_153808 | 1.03 | −1.30 | 2.67 |
| 1423770_at | 217353 | Tmc6 | transmembrane channel-like gene family 6 | NM_145439 /// NM_181321 | 1.23 | −1.11 | 2.56 |
| 1424581_at | 217154 | Stac2 | SH3 and cysteine rich domain 2 | NM_146028 | 1.06 | −1.08 | 2.50 |
| 1425050_at | 66307 | Isoc1 | isochorismatase domain containing 1 | NM_025478 | −1.06 | −1.60 | 2.48 |
| 1435477_s_at | 14130 | Fcgr2b | Fc receptor, IgG, low affinity IIb | NM_001077189 /// NM_010187 | 1.67 | −1.12 | 2.48 |
| 1417933_at | 16012 | Igfbp6 | insulin-like growth factor binding protein 6 | NM_008344 | −1.31 | −1.27 | 2.43 |
| 1437226_x_at | 17357 | Marcksl1 | MARCKS-like 1 | NM_010807 | 1.37 | −1.16 | 2.39 |
| 1419517_at | 72978 | Cnih3 | cornichon homolog 3 (Drosophila) | NM_001160211 /// NM_001160212 /// NM_028408 | 1.23 | −1.10 | 2.38 |
| 1424767_at | 104010 | Cdh22 | cadherin 22 | NM_174988 | 1.57 | −1.08 | 2.36 |
| 1427115_at | 17883 | Myh3 | myosin, heavy polypeptide 3, skeletal muscle, embryonic | NM_001099635 | 1.05 | −1.04 | 2.31 |
| 1416125_at | 14229 | Fkbp5 | FK506 binding protein 5 | NM_010220 | 1.10 | −1.35 | 2.29 |
| 1460366_at | 225898 | Eml3 | echinoderm microtubule associated protein like 3 | NM_144872 | −1.07 | −1.23 | 2.28 |
| 1422552_at | 67874 | Rprm | reprimo, TP53 dependent G2 arrest mediator candidate | NM_023396 | 1.50 | −1.44 | 2.23 |
| 1424451_at | 235674 | Acaa1b | acetyl-Coenzyme A acyltransferase 1B | NM_146230 | 1.08 | 1.10 | 2.21 |
| 1422659_at | 108058 | Camk2d | calcium/calmodulin-dependent protein kinase II, delta | NM_001025438 /// NM_001025439 /// NM_023813 | 1.53 | −1.30 | 2.20 |
| 1419470_at | 14696 | Gnb4 | guanine nucleotide binding protein (G protein), beta 4 | NM_013531 | 1.20 | −1.03 | 2.18 |
| 1448807_at | 99296 | Hrh3 | histamine receptor H3 | NM_133849 /// NR_102309 | 1.00 | −1.18 | 2.17 |
| 1449839_at | 12367 | Casp3 | caspase 3 | NM_009810 | 1.93 | 1.51 | 2.16 |
| 1421505_at | 27217 | Mixl1 | Mix1 homeobox-like 1 (Xenopus laevis) | NM_013729 | 1.03 | −1.54 | 2.15 |
| 1423551_at | 12554 | Cdh13 | cadherin 13 | NM_019707 | −1.31 | −1.22 | 2.13 |
| 1449837_at | 66712 | Spesp1 | sperm equatorial segment protein 1 | NM_025721 | −1.14 | −1.72 | 2.10 |
| 1419606_a_at | 21955 | Tnnt1 | troponin T1, skeletal, slow | NM_001277903 /// NM_001277904 /// NM_011618 | −1.02 | −1.47 | 2.07 |
| 1438673_at | 218756 | Slc4a7 | solute carrier family 4, sodium bicarbonate cotransporter, member 7 | NM_001033270 | 1.61 | 1.01 | 2.07 |
| 1424902_at | 72324 | Plxdc1 | plexin domain containing 1 | NM_001163608 /// NM_028199 | −1.31 | −1.14 | 2.06 |
| 1448635_at | 14211 | Smc2 | structural maintenance of chromosomes 2 | NM_008017 | −1.45 | −1.57 | 2.05 |
| 1426936_at | 192885 /// 215866 /// 629242 /// 641366 | BC005512 /// F630007L15Rik /// Gm6958 /// LOC215866 | cDNA sequence BC005512 /// RIKEN cDNA F630007L15 gene /// predicted gene 6958 /// uncha | XM_001479180 /// XM_001480210 /// XM_003688870 /// XM_003689442 /// XM_989008 /// XR_14 | 1.65 | 1.11 | 2.04 |
| 1450998_at | 65020 | Zfp110 | zinc finger protein 110 | NM_022981 | 1.64 | 1.08 | 2.04 |
| 1448823_at | 20315 | Cxcl12 | chemokine (C-X-C motif) ligand 12 | NM_001012477 /// NM_013655 /// NM_021704 | −1.41 | −1.33 | −2.76 |
| 1418476_at | 12931 | Crlf1 | cytokine receptor-like factor 1 | NM_018827 | −1.12 | −1.05 | −2.77 |
| 1427351_s_at | 16019 | Ighm | immunoglobulin heavy constant mu | --- | 1.08 | 1.42 | −3.06 |
| Overlap (222 genes) | |||||||
|---|---|---|---|---|---|---|---|
| Probeset ID | Entrez Gene |
Gene Symbol |
Gene Title | RefSeq Transcript ID |
WI vs WS |
TI vs TS |
TS vs WS |
| 1450716_at | 11504 | Adamts1 | A disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 mo | NM_009621 | 4.0 | 5.0 | 1.0 |
| 1419706_a_at | 83397 | Akap12 | A kinase (PRKA) anchor protein (gravin) 12 | NM_031185 | 3.5 | 3.8 | −1.0 |
| 1449363_at | 11910 | Atf3 | Activating transcription factor 3 | NM_007498 | 46.7 | 33.1 | −1.2 |
| 1432007_s_at | 11772 | Ap2a2 | Adaptor-related protein complex 2, alpha 2 subunit | NM_007459 | 2.3 | 2.2 | 1.2 |
| 1418823_at | 11845 | Arf6 | ADP-ribosylation factor 6 | NM_007481 | 2.5 | 2.0 | 1.0 |
| 1418250_at | 80981 | Arl4d | ADP-ribosylation factor-like 4D | NM_025404 | 4.5 | 2.6 | 1.1 |
| 1423420_at | 11554 | Adrb1 | Adrenergic receptor, beta 1 | NM_007419 | 3.1 | 2.3 | 1.0 |
| 1416077_at | 11535 | Adm | Adrenomedullin | NM_009627 | 2.2 | 3.0 | −1.1 |
| 1417130_s_at | 57875 | Angptl4 | Angiopoietin-like 4 | NM_020581 | 4.2 | 2.7 | 1.4 |
| 1453287_at | 67434 | Ankrd33b | Ankyrin repeat domain 33B | NM_001164441 /// NM_026153/// NM_027496 | 2.7 | 2.2 | −1.1 |
| 1419091_a_at | 12306 | Anxa2 | Annexin A2 | NM_007585 | 2.8 | 2.5 | −1.1 |
| 1424481_s_at | 494468 | Armcx5 | Armadillo repeat containing, X-linked 5 | NM_001009575 | 2.5 | 2.1 | 1.2 |
| 1451340_at | 214855 | Arid5a | AT rich interactive domain 5A (MRF1-like) | NM_001172205 /// NM_001172206 /// NM_145996 /// NR_033310 | 3.3 | 4.1 | −1.3 |
| 1420973_at | 71371 | Arid5b | AT rich interactive domain 5B (MRF1-like) | NM_023598 | 2.7 | 2.2 | 1.1 |
| 1419004_s_at | 12044/// 12045/// 12047 | Bcl2a1a /// Bcl2a1b /// Bcl2a1d | B cell leukemia/lymphoma 2 related protein A1a /// B cell leukemia/lymphoma 2 related p | NM_007534 /// NM_007536 /// NM_009742 | 3.1 | 2.6 | −1.1 |
| 1416250_at | 12227 | Btg2 | B cell translocation gene 2, anti-proliferative | NM_007570 | 4.2 | 3.2 | 1.2 |
| 1422452_at | 29810 | Bag3 | BCL2-associated athanogene 3 | NM_013863 | 5.5 | 3.0 | 1.2 |
| 1423753_at | 68010 | Bambi | BMP and activin membrane-bound inhibitor | NM_026505 | 2.1 | 2.2 | 1.5 |
| 1437419_at | 140780 | Bmp2k | BMP2 inducible kinase | NM_080708 | 3.7 | 2.2 | 1.4 |
| 1422169_a_at | 12064 | Bdnf | Brain derived neurotrophic factor | NM_001048139 /// NM_001048141 /// NM_001048142 /// NM_007540 | 2.3 | 2.3 | −1.4 |
| 1433956_at | 12562 | Cdh5 | Cadherin 5 | NM_009868 | 2.6 | 2.5 | −1.1 |
| 1437270_a_at | 56708 | Clcf1 | Cardiotrophin-like cytokine factor 1 | NM_019952 | 2.4 | 2.1 | 1.0 |
| 1427844_a_at | 12608 | Cebpb | CCAAT/enhancer binding protein (C/EBP), beta | NM_009883 | 2.9 | 2.2 | −1.1 |
| 1417268_at | 12475 | Cd14 | CD14 antigen | NM_009841 | 5.1 | 4.2 | −1.1 |
| 1416034_at | 12484 | Cd24a | CD24a antigen | NM_009846 | 2.8 | 2.1 | 2.1 |
| 1423760_at | 12505 | Cd44 | CD44 antigen | NM_001039150 /// NM_001039151 /// NM_001177785 /// NM_001177786 /// NM_001177787 /// NM | 6.2 | 4.4 | −1.0 |
| 1419589_at | 17064 | Cd93 | CD93 antigen | NM_010740 | 3.0 | 3.0 | −1.2 |
| 1450842_a_at | 12615 | Cenpa | Centromere protein A | NM_007681 | 6.2 | 5.2 | −1.2 |
| 1427205_x_at | 76380 | Cep112 | Centrosomal protein 112 | NM_029586 /// NM_029606 /// NM_145688 | 2.0 | 2.1 | 1.0 |
| 1451382_at | 69065 | Chac1 | ChaC, cation transport regulator 1 | NM_026929 | 7.9 | 5.9 | 1.0 |
| 1419561_at | 20302 | Ccl3 | Chemokine (C-C motif) ligand 3 | NM_011337 | 4.5 | 4.6 | −1.3 |
| 1419209_at | 14825 | Cxcl1 | Chemokine (C-X-C motif) ligand 1 | NM_008176 | 9.4 | 8.4 | 1.0 |
| 1424143_a_at | 67177 | Cdt1 | Chromatin licensing and DNA replication factor 1 | NM_026014 | 2.1 | 2.1 | 1.1 |
| 1431166_at | 12648 | Chd1 | Chromodomain helicase DNA binding protein 1 | NM_007690 | 2.2 | 2.2 | 1.0 |
| 1424409_at | 71908 | Cldn23 | Claudin 23 | NM_027998 | 2.2 | 2.1 | −1.0 |
| 1452414_s_at | 108673 | Ccdc86 | Coiled-coil domain containing 86 | NM_023731 | 4.8 | 3.5 | 1.2 |
| 1452035_at | 12826 | Col4a1 | Collagen, type IV, alpha 1 | NM_009931 | 2.8 | 2.2 | 1.3 |
| 1419483_at | 12267 | C3ar1 | Complement component 3a receptor 1 | NM_009779 | 3.0 | 2.5 | −1.1 |
| 1416953_at | 14219 | Ctgf | Connective tissue growth factor | NM_010217 | 3.5 | 3.7 | −1.2 |
| 1434618_at | 233490 | Crebzf | CREB/ATF bZIP transcription factor | NM_145151 /// NR_073436 /// NR_073437 /// NR_073438 | −2.8 | −2.5 | −1.1 |
| 1433733_a_at | 12952 | Cry1 | Cryptochrome 1 (photolyase-like) | NM_007771 | 2.7 | 2.4 | −1.0 |
| 1423622_a_at | 56706 | Ccnl1 | Cyclin L1 | NM_001025442 /// NM_001025443 /// NM_019937 | 2.3 | 2.1 | −1.1 |
| 1421679_a_at | 12575 | Cdkn1a | Cyclin-dependent kinase inhibitor 1A (P21) | NM_001111099 /// NM_007669 | 4.1 | 3.0 | 1.2 |
| 1438133_a_at | 16007 | Cyr61 | Cysteine rich protein 61 | NM_010516 | 14.5 | 21.3 | 1.1 |
| 1422533_at | 13121 | Cyp51 | Cytochrome P450, family 51 | NM_020010 | 2.8 | 2.1 | 1.1 |
| 1455372_at | 208922 | Cpeb3 | Cytoplasmic polyadenylation element binding protein 3 | NM_198300 | 2.6 | 2.0 | −1.0 |
| 1449931_at | 67579 | Cpeb4 | Cytoplasmic polyadenylation element binding protein 4 | NM_026252 | 2.8 | 2.1 | 1.2 |
| 1416814_at | 21841 | Tia1 | Cytotoxic granule-associated RNA binding protein 1 | NM_001164078 /// NM_001164079 /// NM_011585 | −2.2 | −2.1 | 1.1 |
| 1448471_a_at | 13024 | Ctla2a | Cytotoxic T lymphocyte-associated protein 2 alpha | NM_001145799 /// NM_007796 | 4.4 | 3.6 | −1.2 |
| 1452352_at | 13025 | Ctla2b | Cytotoxic T lymphocyte-associated protein 2 beta | NM_001145801 /// NM_007797 | 2.6 | 2.5 | −1.1 |
| 1417937_at | 59036 | Dact1 | Dapper homolog 1, antagonist of beta-catenin (xenopus) | NM_001190466 /// NM_021532 | 2.5 | 3.2 | 1.2 |
| 1426081_a_at | 13371 | Dio2 | Deiodinase, iodothyronine, type II | NM_010050 | 3.0 | 2.8 | 1.1 |
| 1417516_at | 13198 | Ddit3 | DNA-damage inducible transcript 3 | NM_007837 | 2.4 | 2.1 | 1.0 |
| 1416756_at | 81489 | Dnajb1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | NM_018808 | 5.5 | 5.7 | −1.4 |
| 1448830_at | 19252 | Dusp1 | Dual specificity phosphatase 1 | NM_013642 | 2.7 | 4.0 | −1.2 |
| 1418401_a_at | 70686 | Dusp16 | Dual specificity phosphatase 16 | NM_001048054 /// NM_130447/// NM_181320 | 3.3 | 3.1 | −1.0 |
| 1415834_at | 67603 | Dusp6 | Dual specificity phosphatase 6 | NM_026268 | 2.1 | 3.0 | −1.7 |
| 1417065_at | 13653 | Egr1 | Early growth response 1 | NM_007913 | 2.6 | 4.3 | −2.0 |
| 1427683_at | 13654 | Egr2 | Early growth response 2 | NM_010118 | 17.4 | 22.0 | −2.3 |
| 1449977_at | 13656 | Egr4 | Early growth response 4 | NM_020596 | 2.3 | 2.2 | −1.4 |
| 1416529_at | 13730 | Emp1 | Epithelial membrane protein 1 | NM_010128 | 7.4 | 5.7 | 1.1 |
| 1416129_at | 74155 | Errfi1 | ERBB receptor feedback inhibitor 1 | NM_133753 | 3.0 | 2.0 | −1.0 |
| 1418294_at | 54357 | Epb4.1l4b | Erythrocyte protein band 4.1-like 4b | NM_019427 | 2.5 | 2.6 | 1.0 |
| 1418635_at | 27049 | Etv3 | ets variant gene 3 | NM_001083318 /// NM_012051 | 4.3 | 2.3 | 1.0 |
| 1441023_at | 67204 | Eif2s2 | Eukaryotic translation initiation factor 2, subunit 2 (beta) | NM_026030 | 2.7 | 2.1 | −1.0 |
| 1417562_at | 13685 | Eif4ebp1 | Eukaryotic translation initiation factor 4E binding protein 1 | NM_007918 | 3.8 | 2.9 | 1.2 |
| 1438427_at | 67544 | Fam120b | Family with sequence similarity 120, member B | NM_024203 /// NR_033586 | 3.4 | 2.4 | 1.3 |
| 1423100_at | 14281 | Fos | FBJ osteosarcoma oncogene | NM_010234 | 11.4 | 20.2 | −1.8 |
| 1422134_at | 14282 | Fosb | FBJ osteosarcoma oncogene B | NM_008036 | 8.9 | 6.6 | −1.1 |
| 1451264_at | 319710 | Frmd6 | FERM domain containing 6 | NM_028127 | 4.3 | 3.2 | −1.2 |
| 1418534_at | 57265 | Fzd2 | frizzled homolog 2 (Drosophila) | NM_020510 | −2.5 | −2.3 | 1.1 |
| 1450135_at | 14365 | Fzd3 | frizzled homolog 3 (Drosophila) | NM_021458 | 3.2 | 2.7 | 1.0 |
| 1419301_at | 14366 | Fzd4 | frizzled homolog 4 (Drosophila) | NM_008055 | 2.9 | 3.0 | −1.0 |
| 1419322_at | 13998 | Fgd6 | FYVE, RhoGEF and PH domain containing 6 | NM_053072 | 2.7 | 2.5 | 1.1 |
| 1422542_at | 23890 | Gpr34 | G protein-coupled receptor 34 | NM_011823 | −2.5 | −2.7 | 1.1 |
| 1460275_at | 14748 | Gpr3 | G-protein coupled receptor 3 | NM_008154 | 2.5 | 2.0 | 1.0 |
| 1420394_s_at | 14727 /// 14728 | Gp49a /// Lilrb4 | Glycoprotein 49 A /// leukocyte immunoglobulin-like receptor, subfamily B, member 4 | NM_008147 /// NM_013532 | 4.1 | 2.9 | 1.0 |
| 1424825_a_at | 14663 | Glycam1 | Glycosylation dependent cell adhesion molecule 1 | NM_008134 | 2.5 | 2.0 | −1.0 |
| 1449519_at | 13197 | Gadd45a | Growth arrest and DNA-damage-inducible 45 alpha | NM_007836 | 4.4 | 3.7 | −1.2 |
| 1449773_s_at | 17873 | Gadd45b | Growth arrest and DNA-damage-inducible 45 beta | NM_008655 | 11.7 | 6.5 | 1.0 |
| 1453851_a_at | 23882 | Gadd45g | Growth arrest and DNA-damage-inducible 45 gamma | NM_011817 | 10.8 | 6.5 | 1.1 |
| 1426063_a_at | 14579 | Gem | GTP binding protein (gene overexpressed in skeletal muscle) | NM_010276 | 8.3 | 10.1 | 1.0 |
| 1423143_at | 69237 | Gtpbp4 | GTP binding protein 4 | NM_027000 | 2.9 | 2.2 | 1.1 |
| 1420499_at | 14528 | Gch1 | GTP cyclohydrolase 1 | NM_008102 | 2.3 | 2.0 | −1.4 |
| 1430295_at | 14674 | Gna13 | Guanine nucleotide binding protein, alpha 13 | NM_010303 | 2.0 | 2.1 | 1.1 |
| 1423566_a_at | 15505 | Hsph1 | Heat shock 105kDa/110kDa protein 1 | NM_013559 | 3.8 | 3.2 | −1.2 |
| 1425964_x_at | 15507 | Hspb1 | Heat shock protein 1 | NM_013560 | 10.7 | 15.2 | −1.2 |
| 1422579_at | 15528 | Hspe1 | Heat shock protein 1 (chaperonin 10) | NM_008303 | 2.6 | 2.1 | 1.0 |
| 1452388_at | 193740 | Hspa1a | Heat shock protein 1A | NM_010479 | 13.9 | 17.6 | −1.4 |
| 1427126_at | 15511 | Hspa1b | Heat shock protein 1B | NM_010478 | 12.9 | 17.6 | −1.5 |
| 1449872_at | 56534 | Hspb3 | Heat shock protein 3 | NM_019960 | 2.7 | 2.2 | −1.0 |
| 1431182_at | 15481 | Hspa8 | Heat shock protein 8 | NM_031165 | 4.4 | 4.6 | −1.4 |
| 1425378_at | 235439 | Herc1 | Hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like do | NM_145617 | 3.9 | 3.2 | 1.3 |
| 1417541_at | 15201 | Hells | Helicase, lymphoid specific | NM_008234 | 2.7 | 2.4 | 1.3 |
| 1448239_at | 15368 | Hmox1 | Heme oxygenase (decycling) 1 | NM_010442 | 7.1 | 5.3 | 1.3 |
| 1418350_at | 15200 | Hbegf | Heparin-binding EGF-like growth factor | NM_010415 | 5.5 | 3.2 | 1.4 |
| 1452534_a_at | 97165 | Hmgb2 | High mobility group box 2 | NM_008252 | 3.2 | 3.4 | 1.0 |
| 1435866_s_at | 319162 | Hist3h2a | Histone cluster 3, H2a | NM_178218 | −2.0 | −2.6 | 1.3 |
| 1419905_s_at | 15446 | Hpgd | Hydroxyprostaglandin dehydrogenase 15 (NAD) | NM_008278 | −2.7 | −2.2 | 1.3 |
| 1416442_at | 15936 | Ier2 | Immediate early response 2 | NM_010499 | 7.4 | 6.6 | −1.0 |
| 1417612_at | 15939 | Ier5 | Immediate early response 5 | NM_010500 | 3.8 | 2.8 | −1.4 |
| 1437103_at | 319765 | Igf2bp2 | Insulin-like growth factor 2 mRNA binding protein 2 | NM_183029 | 3.3 | 2.0 | −1.1 |
| 1424067_at | 15894 | Icam1 | Intercellular adhesion molecule 1 | NM_010493 | 2.2 | 2.3 | −1.3 |
| 1416067_at | 15982 | Ifrd1 | Interferon-related developmental regulator 1 | NM_013562 | 3.7 | 2.4 | 1.1 |
| 1426597_s_at | 212632 | Iffo2 | Intermediate filament family orphan 2 | NM_001205173 /// NM_183148 | 2.5 | 2.0 | 1.1 |
| 1417409_at | 16476 | Jun | Jun oncogene | NM_010591 | 5.8 | 4.2 | 1.4 |
| 1449117_at | 16478 | Jund | Jun proto-oncogene related gene d | NM_010592 | 2.1 | 2.1 | 1.1 |
| 1415899_at | 16477 | Junb | Jun-B oncogene | NM_008416 | 3.2 | 4.1 | −1.7 |
| 1425270_at | 16561 | Kif1b | kinesin family member 1B | NM_008441 /// NM_207682 | 2.5 | 2.6 | −1.2 |
| 1416029_at | 21847 | Klf10 | Kruppel-like factor 10 | NM_013692 | 3.1 | 2.6 | −1.4 |
| 1417394_at | 16600 | Klf4 | Kruppel-like factor 4 (gut) | NM_010637 | 2.4 | 5.9 | 1.0 |
| 1418280_at | 23849 | Klf6 | Kruppel-like factor 6 | NM_011803 | 4.7 | 5.5 | 1.1 |
| 1439479_at | 226413 | Lct | Lactase | NM_001081078 | −2.5 | −2.2 | −1.0 |
| 1421654_a_at | 16905 | Lmna | Lamin A | NM_001002011 /// NM_001111102 /// NM_019390 | 5.1 | 4.0 | 1.2 |
| 1426808_at | 16854 | Lgals3 | Lectin, galactose binding, soluble 3 | NM_001145953 /// NM_010705 | 4.7 | 3.9 | −1.1 |
| 1433842_at | 16978 | Lrrfip1 | Leucine rich repeat (in FLII) interacting protein 1 | NM_001111311 /// NM_001111312 /// NM_008515 | 6.6 | 5.2 | 1.0 |
| 1422725_at | 17152 | Mak | Male germ cell-associated kinase | NM_001145802 /// NM_001145803 /// NM_008547 | 3.3 | 4.1 | −1.0 |
| 1429170_a_at | 17764 | Mtf1 | Metal response element binding transcription factor 1 | NM_008636 | 4.5 | 4.8 | 1.1 |
| 1451612_at | 17748 | Mt1 | Metallothionein 1 | NM_013602 | 2.8 | 3.0 | 1.0 |
| 1419254_at | 17768 | Mthfd2 | Methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cycl | NM_008638 | 3.8 | 2.8 | 1.0 |
| 1434364_at | 53859 | Map3k14 | Mitogen-activated protein kinase kinase kinase 14 | NM_016896 | 2.4 | 2.2 | 1.0 |
| 1424942_a_at | 17869 | Myc | Myelocytomatosis oncogene | NM_001177352 /// NM_001177353 /// NM_001177354 /// NM_010849 | 4.0 | 3.8 | 1.1 |
| 1448503_at | 17210 | Mcl1 | Myeloid cell leukemia sequence 1 | NM_008562 | 6.1 | 4.8 | 1.1 |
| 1418589_a_at | 17349 | Mlf1 | Myeloid leukemia factor 1 | NM_001039543 /// NM_010801 | 3.4 | 3.1 | −1.1 |
| 1422818_at | 18003 | Nedd9 | Neural precursor cell expressed, developmentally down-regulated gene 9 | NM_001111324 /// NM_017464 | 3.6 | 3.8 | 1.0 |
| 1420720_at | 53324 | Nptx2 | Neuronal pentraxin 2 | NM_016789 | 7.2 | 3.0 | 1.0 |
| 1417930_at | 17937 | Nab2 | Ngfi-A binding protein 2 | NM_001122895 /// NM_008668 | 2.9 | 2.3 | −1.2 |
| 1417483_at | 80859 | Nfkbiz | Nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta | NM_001159394 /// NM_001159395 /// NM_030612 | 6.4 | 4.6 | 1.4 |
| 1418932_at | 18030 | Nfil3 | Nuclear factor, interleukin 3, regulated | NM_017373 | 5.9 | 7.1 | −1.4 |
| 1428083_at | 66961 | Neat1 | Nuclear paraspeckle assembly transcript 1 (non-protein coding) | NR_003513 | 2.2 | 2.5 | −1.2 |
| 1416505_at | 15370 | Nr4a1 | Nuclear receptor subfamily 4, group A, member 1 | NM_010444 | 2.1 | 3.6 | −2.4 |
| 1436780_at | 108155 | Ogt | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide- | NM_139144 | −2.3 | −2.2 | 1.2 |
| 1418674_at | 18414 | Osmr | Oncostatin M receptor | NM_011019 | 7.2 | 4.1 | 1.3 |
| 1423174_a_at | 58220 | Pard6b | par-6 (partitioning defective 6) homolog beta (C. elegans) | NM_021409 | 3.4 | 2.6 | −1.1 |
| 1422324_a_at | 19227 | Pthlh | Parathyroid hormone-like peptide | NM_008970 | 3.5 | 4.0 | −1.1 |
| 1417372_a_at | 67245 | Peli1 | Pellino 1 | NM_023324 | 2.2 | 2.2 | 1.2 |
| 1419006_s_at | 93834 | Peli2 | Pellino 2 | NM_033602 | 2.1 | 2.1 | −1.0 |
| 1449851_at | 18626 | Per1 | Period circadian clock 1 | NM_001159367 /// NM_011065 | 2.5 | 2.3 | 1.1 |
| 1417602_at | 18627 | Per2 | Period circadian clock 2 | NM_011066 | 2.5 | 2.8 | −1.6 |
| 1420715_a_at | 19016 | Pparg | Peroxisome proliferator activated receptor gamma | NM_001127330 /// NM_011146 | 3.4 | 2.1 | 1.0 |
| 1421811_at | 21825 | Thbs1 | Thrombospondin 1 | NM_011580/// NM_013753 | 13.7 | 8.0 | 1.3 |
| 1421917_at | 18595 | Pdgfra | Platelet derived growth factor receptor, alpha polypeptide | NM_001083316 /// NM_011058 | −2.0 | −2.0 | 1.1 |
| 1418835_at | 21664 | Phlda1 | Pleckstrin homology-like domain, family A, member 1 | NM_009344 | 3.4 | 3.4 | −1.2 |
| 1434496_at | 12795 | Plk3 | Polo-like kinase 3 | NM_013807 | 2.2 | 2.4 | −1.0 |
| 1429456_a_at | 26939 | Polr3e | Polymerase (RNA) III (DNA directed) polypeptide E | NM_001164096 /// NM_025298 | 2.5 | 2.4 | 1.4 |
| 1424874_a_at | 19205 | Ptbp1 | Polypyrimidine tract binding protein 1 | NM_001077363 /// NM_008956 | 2.6 | 2.0 | 1.1 |
| 1425341_at | 16527 | Kcnk3 | Potassium channel, subfamily K, member 3 | NM_010608 | 3.1 | 2.1 | −1.2 |
| 1431213_a_at | 67527 /// 100041932 | Gm3579 /// LOC67527 | Predicted gene 3579 /// murine leukemia retrovirus | XM_001477842 | 2.5 | 2.1 | −1.2 |
| 1433668_at | 108767 | Pnrc1 | Proline-rich nuclear receptor coactivator 1 | NM_001033225 | −2.1 | −2.1 | −1.0 |
| 1424208_at | 19219 | Ptger4 | Prostaglandin E receptor 4 (subtype EP4) | NM_001136079 /// NM_008965 | 4.6 | 2.6 | 1.4 |
| 1417262_at | 19225 | Ptgs2 | Prostaglandin-endoperoxide synthase 2 | NM_011198 | 8.5 | 3.3 | −1.3 |
| 1448325_at | 17872 | Ppp1r15a | Protein phosphatase 1, regulatory (inhibitor) subunit 15A | NM_008654 | 4.2 | 4.7 | 1.0 |
| 1429715_at | 71978 | Ppp2r2a | Protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform | NM_001205188 /// NM_028032 | 2.8 | 2.1 | 1.1 |
| 1449249_at | 54216 | Pcdh7 | Protocadherin 7 | NM_001122758 /// NM_018764 | 3.0 | 2.7 | −1.2 |
| 1435458_at | 18712 | Pim1 | Proviral integration site 1 | NM_008842 | 5.6 | 2.5 | −1.2 |
| 1431724_a_at | 70839 | P2ry12 | Purinergic receptor P2Y, G-protein coupled 12 | NM_027571 | −2.8 | −2.4 | 1.1 |
| 1427931_s_at | 216134 | Pdxk | Pyridoxal (pyridoxine, vitamin B6) kinase | NM_172134 | 2.4 | 2.0 | 1.1 |
| 1417273_at | 27273 | Pdk4 | Pyruvate dehydrogenase kinase, isoenzyme 4 | NM_013743 | 3.0 | 2.2 | 1.1 |
| 1426452_a_at | 75985 | Rab30 | RAB30, member RAS oncogene family | NM_029494 | 2.3 | 2.1 | −1.1 |
| 1448414_at | 19355 | Rad1 | RAD1 homolog (S. pombe) | NM_011232 | −2.1 | −2.1 | 1.0 |
| 1418892_at | 80837 | Rhoj | ras homolog gene family, member J | NM_023275 | 3.4 | 2.2 | 1.3 |
| 1423854_a_at | 68939 | Rasl11b | RAS-like, family 11, member B | NM_026878 | 2.0 | 2.6 | −1.6 |
| 1422562_at | 56437 | Rrad | Ras-related associated with diabetes | NM_019662 | 14.6 | 4.5 | 1.8 |
| 1426037_a_at | 19734 | Rgs16 | regulator of G-protein signaling 16 | NM_011267 | 2.3 | 2.0 | 1.3 |
| 1452359_at | 100532 | Rell1 | RELT-like 1 | NM_145923 | 2.4 | 2.2 | 1.1 |
| 1420710_at | 19696 | Rel | Reticuloendotheliosis oncogene | NM_009044 | 3.4 | 2.6 | −1.1 |
| 1416700_at | 74194 | Rnd3 | Rho family GTPase 3 | NM_028810 | 3.2 | 3.3 | −1.0 |
| 1438502_x_at | 20068 | Rps17 | Ribosomal protein S17 | NM_009092 | 3.1 | 2.8 | −1.1 |
| 1427299_at | 110651 | Rps6ka3 | Ribosomal protein S6 kinase polypeptide 3 | NM_148945 | 2.3 | 2.0 | −1.1 |
| 1427932_s_at | 71719 /// 71739 /// 319269 | 1200003I10Rik /// 1200015M12Rik /// A130040M12Rik | RIKEN cDNA 1200003I10 gene /// RIKEN cDNA 1200015M12 gene /// RIKEN cDNA A130040M12 gen | NR_002860 | 3.7 | 3.8 | −1.2 |
| 1451415_at | 69068 | 1810011O10Rik | RIKEN cDNA 1810011O10 gene | NM_026931 | 2.3 | 2.5 | 1.3 |
| 1428552_at | 66520 | 2610001J05Rik | RIKEN cDNA 2610001J05 gene | NM_183258 /// NR_024619 | −2.1 | −2.2 | 1.1 |
| 1416355_at | 19655 | Rbmx | RNA binding motif protein, X chromosome | NM_001166623 /// NM_011252 /// NR_029425 | −2.1 | −2.1 | −1.2 |
| 1424704_at | 12393 | Runx2 | Runt related transcription factor 2 | NM_001145920 /// NM_001146038 /// NM_001271627 /// NM_001271630 /// NM_001271631 /// NM | 2.6 | 2.3 | −1.1 |
| 1419149_at | 18787 | Serpine1 | Serine (or cysteine) peptidase inhibitor, clade E, member 1 | NM_008871 | 3.2 | 3.0 | −1.1 |
| 1429650_at | 74178 | Stk40 | Serine/threonine kinase 40 | NM_001145827 /// NM_028800 | 3.4 | 2.4 | 1.0 |
| 1417406_at | 55942 | Sertad1 | SERTA domain containing 1 | NM_018820 | 2.9 | 2.4 | −1.2 |
| 1425139_at | 230784 | Sesn2 | Sestrin 2 | NM_144907 | 2.7 | 2.3 | 1.0 |
| 1448170_at | 20439 | Siah2 | Seven in absentia 2 | NM_009174 | 2.6 | 2.6 | −1.1 |
| 1433674_a_at | 83673 | Snhg1 | Small nucleolar RNA host gene (non-protein coding) 1 | NR_002896 | 4.6 | 3.6 | 1.2 |
| 1428529_at | 72655 | Snhg5 | Small nucleolar RNA host gene 5 | NR_040721 | 2.6 | 2.2 | 1.1 |
| 1428776_at | 75750 | Slc10a6 | Solute carrier family 10 (sodium/bile acid cotransporter family), member 6 | NM_029415 | 5.4 | 5.4 | −1.2 |
| 1420697_at | 65221 | Slc15a3 | Solute carrier family 15, member 3 | NM_023044 | 2.6 | 2.6 | −1.2 |
| 1438824_at | 20515 | Slc20a1 | Solute carrier family 20, member 1 | NM_001159593 /// NM_015747 | 2.3 | 2.7 | 1.1 |
| 1422786_at | 22782 | Slc30a1 | Solute carrier family 30 (zinc transporter), member 1 | NM_009579 | 2.6 | 2.4 | 1.0 |
| 1420405_at | 28250 | Solute carrier organic anion transporter family, member 1a4 | NM_030687 | −2.6 | −2.3 | −1.1 | |
| 1422256_at | 20606 | Sstr2 | Somatostatin receptor 2 | NM_001042606 /// NM_009217 | 3.4 | 2.8 | −1.1 |
| 1451596_a_at | 20698 | Sphk1 | Sphingosine kinase 1 | NM_001172472 /// NM_001172473 /// NM_001172475 /// NM_011451 /// NM_025367 | 2.5 | 2.7 | −1.3 |
| 1420150_at | 74646 | Spsb1 | splA/ryanodine receptor domain and SOCS box containing 1 | NM_029035 | 2.6 | 2.4 | −1.2 |
| 1436584_at | 24064 | Spry2 | Sprouty homolog 2 (Drosophila) | NM_011897 | 3.5 | 2.4 | −1.2 |
| 1449109_at | 216233 | Socs2 | Suppressor of cytokine signaling 2 | NM_001168655 /// NM_001168656 /// NM_001168657 /// NM_007706 | 2.1 | 2.3 | −1.1 |
| 1456212_x_at | 12702 | Socs3 | Suppressor of cytokine signaling 3 | NM_007707 | 11.6 | 13.1 | −1.3 |
| 1434089_at | 104027 | Synpo | Synaptopodin | NM_001109975 /// NM_177340 | 2.3 | 2.5 | −1.1 |
| 1430271_x_at | 75316 | Taf1d | TATA box binding protein (Tbp)-associated factor, RNA polymerase I, D | NM_026541 /// NM_027261 /// NM_029248 /// NR_028401 | 2.1 | 2.3 | 1.1 |
| 1452161_at | 99929 | Tiparp | TCDD-inducible poly(ADP-ribose) polymerase | NM_178892 | 4.8 | 2.8 | 1.2 |
| 1416342_at | 21923 | Tnc | Tenascin C | NM_011607 | 5.8 | 4.3 | 1.2 |
| 1424012_at | 78802 | Ttc30a1 | Tetratricopeptide repeat domain 30A1 | NM_030188 | −2.4 | −2.1 | 1.0 |
| 1418547_at | 21789 | Tfpi2 | Tissue factor pathway inhibitor 2 | NM_009364 | 2.6 | 2.0 | −1.1 |
| 1454018_at | 24086 | Tlk2 | Tousled-like kinase 2 (Arabidopsis) | NM_001112705 /// NM_011903 | 2.6 | 2.5 | 1.2 |
| 1436854_at | 22064 | Trpc2 | Transient receptor potential cation channel, subfamily C, member 2 | NM_001109897 /// NM_011644 | −2.2 | −2.4 | 1.1 |
| 1423289_a_at | 66282 | Tma16 | Translation machinery associated 16 homolog (S. cerevisiae) | NM_025465 | 2.5 | 2.2 | 1.1 |
| 1424880_at | 211770 | Trib1 | Tribbles homolog 1 (Drosophila) | NM_144549 | 3.9 | 3.8 | −1.3 |
| 1430576_at | 22019 | Tpp2 | Tripeptidyl peptidase II | NM_009418 | 2.5 | 2.4 | 1.2 |
| 1416431_at | 67951 | Tubb6 | Tubulin, beta 6 class V | NM_026473 | 5.9 | 6.4 | −1.0 |
| 1418572_x_at | 27279 | Tnfrsf12a | Tumor necrosis factor receptor superfamily, member 12a | NM_001161746 /// NM_013749 | 7.0 | 6.7 | −1.2 |
| 1456094_at | 72344 | Usp36 | Ubiquitin specific peptidase 36 | NM_001033528 | 3.1 | 2.4 | −1.1 |
| 1452385_at | 99526 | Usp53 | Ubiquitin specific peptidase 53 | NM_133857 | 3.1 | 2.3 | 1.1 |
| 1436882_at | 66177 | Ubl5 | Ubiquitin-like 5 | NM_025401 | 2.1 | 2.1 | 1.2 |
| 1454842_a_at | 97884 | B3galnt2 | UDP-GalNAc:betaGlcNAcbeta 1,3-galactosaminyltransferase, polypeptide 2 | NM_178640 | −2.9 | −3.2 | 1.2 |
| 1420994_at | 108105 | B3gnt5 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 | NM_001159407 /// NM_001159408 /// NM_054052 | 2.5 | 2.8 | −1.2 |
| 1434606_at | 13867 | Erbb3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | NM_010153 | −3.5 | −2.3 | −1.4 |
| 1418936_at | 17133 | Maff | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) | NM_010755 | 6.0 | 5.6 | −1.1 |
| 1455812_x_at | 246154 | Vasn | vasorin | NM_139307 | 2.3 | 2.1 | 1.1 |
| 1456292_a_at | 22352 | Vim | Vimentin | NM_011701 | 2.5 | 2.1 | −1.1 |
| 1434744_at | 230734 | Yrdc | yrdC domain containing (E.coli) | NM_153566 | 2.1 | 2.1 | 1.0 |
| 1422570_at | 22632 | Yy1 | YY1 transcription factor | NM_009537 | 2.2 | 2.1 | 1.0 |
| 1425305_at | 114565 | Zbtb21 | Zinc finger and BTB domain containing 21 | NM_001081684 /// NM_001081685 /// NM_175428 | 2.4 | 2.1 | 1.0 |
| 1452519_a_at | 22695 | Zfp36 | Zinc finger protein 36 | NM_011756 | 4.1 | 5.2 | −1.1 |
| 1424974_at | 232854 | Zfp418 | Zinc finger protein 418 | NM_146179 | −2.3 | −2.3 | −1.1 |
| 1449946_a_at | 68040 | Zfp593 | Zinc finger protein 593 | NM_024215 | 2.5 | 2.4 | −1.1 |
| 1452623_at | 268670 | Zfp759 | Zinc finger protein 759 | NM_172392 | −2.1 | −2.4 | 1.3 |
| 1427539_a_at | 52696 | Zwint | ZW10 interactor | NM_025635 | 4.9 | 4.1 | 1.2 |
| Overlap (1 genes) | |||
|---|---|---|---|
| RefSeq transcript ID | WI vs WS | TI vs TS | TS vs WS |
| NM_001276684 /// NM_018790 | 6.15 | 8.25 | −2.46 |
| Overlap (2 genes) | |||||
|---|---|---|---|---|---|
| Gene symbol |
Gene title | RefSeq transcript ID | WI vs WS | TI vs TS | TS vs WS |
| Sox11 | SRY-box containing gene 11 | NM_009234 | 3.51 | −1.37 | 3.32 |
| Upp1 | uridine phosphorylase 1 | NM_001159401 /// NM_001159402 /// NM_009477 | 2.14 | 1.04 | 2.14 |
| Overlap (3 genes) | |||||||
|---|---|---|---|---|---|---|---|
| Probeset ID | Entrez gene |
Gene symbol |
gene title | RefSeq transcript ID | WI vs WS |
TI vs TS |
TS vs WS |
| 1418160_at | 22652 | Mkrn3 | makorin, ring finger protein, 3 | NM_011746 | −1.47 | −2.05 | 5.40 |
| 1418475_at | 20277 | Scnn1b | sodium channel, nonvoltage-gated 1 beta | NM_001272023 /// NM_011325 /// NR_073548 | −1.20 | −2.61 | 3.66 |
| 1419663_at | 18295 | Ogn | osteoglycin | NM_008760 | −1.30 | −2.81 | 5.70 |
Footnotes
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
REFERENCES
- Buttner F, Cordes C, Gerlach F, Heimann A, Alessandri B, Luxemburger U, Tureci O, Hankeln T, Kempski O, Burmester T. Genomic response of the rat brain to global ischemia and reperfusion. Brain Res. 2009;1252:1–14. doi: 10.1016/j.brainres.2008.10.045. [DOI] [PubMed] [Google Scholar]
- Chen J, Uchimura K, Stetler RA, Zhu RL, Nakayama M, Jin K, Graham SH, Simon RP. Transient global ischemia triggers expression of the DNA damage-inducible gene GADD45 in the rat brain. J Cereb Blood Flow Metab. 1998;18:646–657. doi: 10.1097/00004647-199806000-00007. [DOI] [PubMed] [Google Scholar]
- Chymkowitch P, Nguea PA, Enserink JM. SUMO-regulated transcription: challenging the dogma. BioEssays. 2015;37:1095–1105. doi: 10.1002/bies.201500065. [DOI] [PubMed] [Google Scholar]
- Datwyler AL, Lattig-Tunnemann G, Yang W, Paschen W, Lee SL, Dirnagl U, Endres M, Harms C. SUMO2/3 conjugation is an endogenous neuroprotective mechanism. J Cereb Blood Flow Metab. 2011;31:2152–2159. doi: 10.1038/jcbfm.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- Feng Z, Davis DP, Sasik R, Patel HH, Drummond JC, Patel PM. Pathway and gene ontology based analysis of gene expression in a rat model of cerebral ischemic tolerance. Brain Res. 2007;1177:103–123. doi: 10.1016/j.brainres.2007.07.047. [DOI] [PubMed] [Google Scholar]
- Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- Jin K, Mao XO, Eshoo MW, Nagayama T, Minami M, Simon RP, Greenberg DA. Microarray analysis of hippocampal gene expression in global cerebral ischemia. Ann Neurol. 2001;50:93–103. doi: 10.1002/ana.1073. [DOI] [PubMed] [Google Scholar]
- Kawahara N, Wang Y, Mukasa A, Furuya K, Shimizu T, Hamakubo T, Aburatani H, Kodama T, Kirino T. Genome-wide gene expression analysis for induced ischemic tolerance and delayed neuronal death following transient global ischemia in rats. J Cereb Blood Flow Metab. 2004;24:212–223. doi: 10.1097/01.WCB.0000106012.33322.A2. [DOI] [PubMed] [Google Scholar]
- Krumova P, Weishaupt JH. Sumoylation in neurodegenerative diseases. Cell Mol Life Sci. 2013;70:2123–2138. doi: 10.1007/s00018-012-1158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Hallenbeck JM. SUMO and ischemic tolerance. Neuromolecular Med. 2013;15:771–781. doi: 10.1007/s12017-013-8239-9. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Miyake S, Wakita H, McMullen DC, Azuma Y, Auh S, Hallenbeck JM. Protein SUMOylation is massively increased in hibernation torpor and is critical for the cytoprotection provided by ischemic preconditioning and hypothermia in SHSY5Y cells. J Cereb Blood Flow Metab. 2007;27:950–962. doi: 10.1038/sj.jcbfm.9600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Castri P, Bembry J, Maric D, Auh S, Hallenbeck JM. SUMOylation participates in induction of ischemic tolerance. J Neurochem. 2009;109:257–267. doi: 10.1111/j.1471-4159.2009.05957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Mou Y, Maric D, Klimanis D, Auh S, Hallenbeck JM. Elevated global SUMOylation in Ubc9 transgenic mice protects their brains against focal cerebral ischemic damage. PLoS One. 2011;6:e25852. doi: 10.1371/journal.pone.0025852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Bernstock JD, Nagaraja N, Ko B, Hallenbeck JM. Global SUMOylation facilitates the multimodal neuroprotection afforded by quercetin against the deleterious effects of oxygen/glucose deprivation and the restoration of oxygen/glucose. J Neurochem. 2016;138:101–116. doi: 10.1111/jnc.13643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Bei D, Parmar H, Matus A. Activity-regulated, cytoskeleton-associated protein (Arc) is essential for visceral endoderm organization during early embryogenesis. Mech Dev. 2000;92:207–215. doi: 10.1016/s0925-4773(99)00340-8. [DOI] [PubMed] [Google Scholar]
- Liu HW, Zhang J, Heine GF, Arora M, Gulcin OH, Onti-Srinivasan R, Huang K, Parvin JD. Chromatin modification by SUMO-1 stimulates the promoters of translation machinery genes. Nucleic Acids Res. 2012;40:10172–10186. doi: 10.1093/nar/gks819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Sheng H, Yu Z, Paschen W, Yang W. O-linked beta-N-acetylglucosamine modification of proteins is activated in post-ischemic brains of young but not aged mice: Implications for impaired functional recovery from ischemic stress. J Cereb Blood Flow Metab. 2016;36:393–398. doi: 10.1177/0271678X15608393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyret-Kahn H, Benhamed M, Ye T, Le GS, Cossec JC, Lapaquette P, Bischof O, Ouspenskaia M, Dasso M, Seeler J, Davidson I, Dejean A. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 2013;23:1563–1579. doi: 10.1101/gr.154872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati A, Graziano V, De LV, Pascale M, Turco MC. BAG3: a multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011;2:e141. doi: 10.1038/cddis.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- Schmidt-Kastner R. Genomic approach to selective vulnerability of the hippocampus in brain ischemia-hypoxia. Neuroscience. 2015;309:259–279. doi: 10.1016/j.neuroscience.2015.08.034. [DOI] [PubMed] [Google Scholar]
- Schmidt-Kastner R, Zhang B, Belayev L, Khoutorova L, Amin R, Busto R, Ginsberg MD. DNA microarray analysis of cortical gene expression during early recirculation after focal brain ischemia in rat. Brain Res Mol Brain Res. 2002;108:81–93. doi: 10.1016/s0169-328x(02)00516-8. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Seifert A, Schofield P, Barton GJ, Hay RT. Proteotoxic stress reprograms the chromatin landscape of SUMO modification. Sci Signal. 2015;8:rs7. doi: 10.1126/scisignal.aaa2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Bear MF. New views of Arc, a master regulator of synaptic plasticity. Nat Neurosci. 2011;14:279–284. doi: 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan FA, Sweatt JD. The role of the Gadd45 family in the nervous system: a focus on neurodevelopment, neuronal injury, and cognitive neuroepigenetics. Adv Exp Med Biol. 2013;793:81–119. doi: 10.1007/978-1-4614-8289-5_6. [DOI] [PubMed] [Google Scholar]
- Wang L, Rodriguiz RM, Wetsel WC, Sheng H, Zhao S, Liu X, Paschen W, Yang W. Neuron-specific Sumo1-3 knockdown in mice impairs episodic and fear memories. J Psychiatry Neurosci. 2014a;39:259–266. doi: 10.1503/jpn.130148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wansleeben C, Zhao S, Miao P, Paschen W, Yang W. SUMO2 is essential while SUMO3 is dispensable for mouse embryonic development. EMBO Rep. 2014b;15:878–885. doi: 10.15252/embr.201438534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellons JC, 3rd, Sheng H, Laskowitz DT, Mackensen GB, Pearlstein RD, Warner DS. A comparison of strain-related susceptibility in two murine recovery models of global cerebral ischemia. Brain Res. 2000;868:14–21. doi: 10.1016/s0006-8993(00)02216-2. [DOI] [PubMed] [Google Scholar]
- Yakubov E, Gottlieb M, Gil S, Dinerman P, Fuchs P, Yavin E. Overexpression of genes in the CA1 hippocampus region of adult rat following episodes of global ischemia. Brain Res Mol Brain Res. 2004;127:10–26. doi: 10.1016/j.molbrainres.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Yang W, Sheng H, Homi HM, Warner DS, Paschen W. Cerebral ischemia/stroke and small ubiquitin-like modifier (SUMO) conjugation–a new target for therapeutic intervention? J Neurochem. 2008a;106:989–999. doi: 10.1111/j.1471-4159.2008.05404.x. [DOI] [PubMed] [Google Scholar]
- Yang W, Sheng H, Warner DS, Paschen W. Transient focal cerebral ischemia induces a dramatic activation of small ubiquitin-like modifier conjugation. J Cereb Blood Flow Metab. 2008b;28:892–896. doi: 10.1038/sj.jcbfm.9600601. [DOI] [PubMed] [Google Scholar]
- Yang W, Sheng H, Warner DS, Paschen W. Transient global cerebral ischemia induces a massive increase in protein sumoylation. J Cereb Blood Flow Metab. 2008c;28:269–279. doi: 10.1038/sj.jcbfm.9600523. [DOI] [PubMed] [Google Scholar]
- Yang W, Ma Q, Mackensen GB, Paschen W. Deep hypothermia markedly activates the small ubiquitin-like modifier conjugation pathway; implications for the fate of cells exposed to transient deep hypothermic cardiopulmonary bypass. J Cereb Blood Flow Metab. 2009;29:886–890. doi: 10.1038/jcbfm.2009.16. [DOI] [PubMed] [Google Scholar]
- Yang W, Thompson JW, Wang Z, Wang L, Sheng H, Foster MW, Moseley MA, Paschen W. Analysis of oxygen/glucose-deprivation-induced changes in SUMO3 conjugation using SILAC-based quantitative proteomics. J Proteome Res. 2012;11:1108–1117. doi: 10.1021/pr200834f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Wang L, Roehn G, Pearlstein RD, Ali-Osman F, Pan H, Goldbrunner R, Krantz M, Harms C, Paschen W. Small ubiquitin-like modifier 1–3 conjugation is activated in human astrocytic brain tumors and is required for glioblastoma cell survival. Cancer Sci. 2013;104:70–77. doi: 10.1111/cas.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Sheng H, Thompson JW, Zhao S, Wang L, Miao P, Liu X, Moseley MA, Paschen W. Small ubiquitin-like modifier 3-modified proteome regulated by brain ischemia in novel small ubiquitin-like modifier transgenic mice: putative protective proteins/pathways. Stroke. 2014;45:1115–1122. doi: 10.1161/STROKEAHA.113.004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Sheng H, Wang H. Targeting the SUMO pathway for neuroprotection in brain ischemia. Stroke Vasc Neurol. 2016;1:101–107. doi: 10.1136/svn-2016-000031. [DOI] [PMC free article] [PubMed] [Google Scholar]