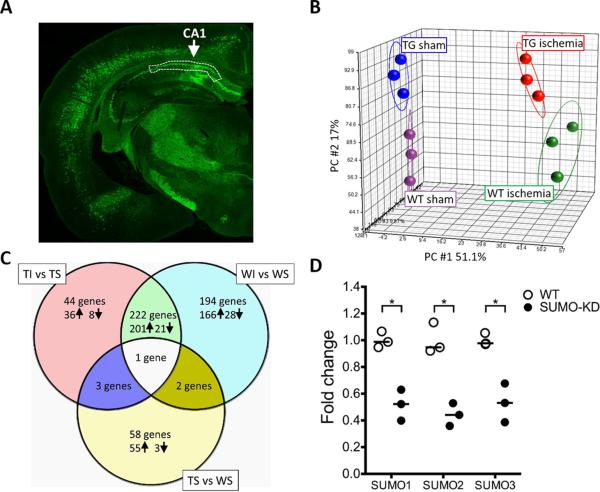
Fig. 2.
Overview of microarray data. Data analysis was performed on the global gene expression profiles of 12 samples from four groups: wild-type (WT) sham (WS), and ischemia (WI); and SUMO-KD (TG) sham (TS), and ischemia (TI). (A) The sampling regions. The region of hippocampal CA1 subfield that was dissected out and used for microarray analysis and qPCR is marked with white dot lines in a representative brain slice of SUMO-KD mice with GFP fluorescence. (B) Principal component analysis (PCA). The individual samples were plotted in a 3-dimensional space based on three principal components. Four groups of samples are clustered according to the genotype and surgery. (C) Venn diagram. The numbers of differentially regulated genes that were identified by pairwise comparisons of groups, with a cut-off of ≥ 2-twofold increase (↑) or decrease (↓) in gene expression are shown. (D) Verification of SUMO1-3 knockdown in SUMO-KD mice. The RNA samples from the sham group that were used for the microarray study, were analyzed to determine the levels of SUMO1-3 mRNA levels in WT and SUMO-KD mice. All individual data were normalized to β-actin. To calculate fold change, the mean values of WT mouse samples were set to 1.0. Horizontal bar = median values; *p ≤ 0.05.