Abstract
Traditionally, biologists have devoted their careers to studying individual biological entities of their own interest, partly due to lack of available data regarding that entity. Large, high-throughput data, too complex for conventional processing methods (i.e., “big data”), has accumulated in cancer biology, which is freely available in public data repositories. Such challenges urge biologists to inspect their biological entities of interest using novel approaches, firstly including repository data retrieval. Essentially, these revolutionary changes demand new interpretations of huge datasets at a systems-level, by so called “systems biology”. One of the representative applications of systems biology is to generate a biological network from high-throughput big data, providing a global map of molecular events associated with specific phenotype changes. In this review, we introduce the repositories of cancer big data and cutting-edge systems biology tools for network generation, and improved identification of therapeutic targets.
Keywords: Cancer, Network biology, Signaling network, Systems biology
INTRODUCTION
Traditionally, researchers have focused their efforts on single biological phenomena (e.g., a single gene mutation) or a specific signaling pathway (1). Now, the age of “omics” big data has brought about cutting-edge processing methods for interpreting biological mega data, which have now universally adopted. Based on such mega data (so-called “big data”), researchers aim to understand systems-level-based phenotype changes (1, 2) by assessing entire pathways/networks, and not just a single entity. Systems biology is defined as a framework (3) to enable systems-level understanding for generating new biological hypotheses, by computational modeling of massive high-throughput data.
Currently, systems biology has broadened its applications from basic science (including small RNAs) (4–6) toward translational medicine, including biomarker and therapeutic target identification (1–3, 7, 8). Systems biology often begins from high-throughput experimental data. Due to mammoth data deposition, as well as data generation by various next-generation sequencing (NGS) techniques (9), big data science has emerged, in particular, from the field of cancer genomics (10). The most widely used repositories include The Cancer Genome Atlas (TCGA) Research Network (11) and the International Cancer Genome Consortium (ICGC) (12). The development of applications for big data science (10) has been facilitated by systems biology frameworks to allow interpretation of systems-level tumorigenesis and molecular mechanisms.
Systems biology covers several diverse areas (13): hypothesis generation and network construction (or inference), and network simulation (e.g., ordinary differential equations, boolean dynamics). In this review, we restrict our discussion to network generation, while also describing analysis tools and relating databases in the field of cancer.
A WORKFLOW OF SYSTEMS BIOLOGY
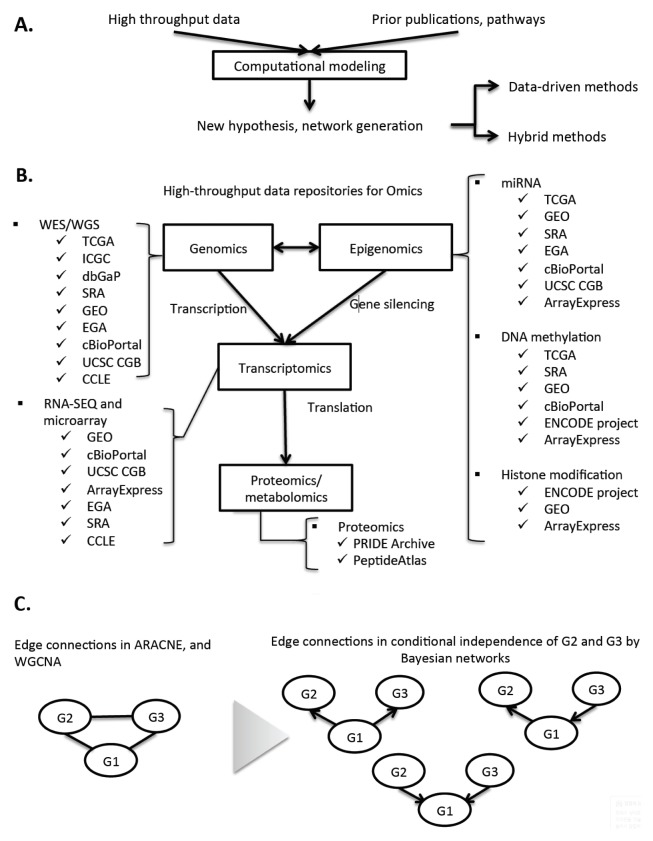
Systems biology has a straightforward workflow of components (13), as shown in Fig. 1A. To understand systems-level biology, observations for all entries are necessary, and high-throughput data is merely a starting point. Computational modeling takes the high-throughput data and, in certain circumstances, prior knowledge (including pathways and gene sets) is selected, resulting in network inference and hypothesis generation (13). Depending on whether computational modeling is used with or without prior knowledge, one may employ both data-driven network modeling and hybrid network modeling, respectively (14). In both of them, computational modeling is a key component, due to its ability to deal with the complexity of interconnectivity among systems entries (13, 14).
Fig. 1.
Systems biology, databases, and network generation. (A) The diversity of types of high-throughput data (genomics, epigenomics, transcriptomics, proteomics, metabolomics) available. The relationships among the data types are connected by edges. (B) The flow (represented by “edges”) of genetic information from DNA to protein is aligned with the diverse data types. Public repositories corresponding to each data type are listed (further description in Table 1). (C) Network differences between correlation-based approaches and Bayesian networks approaches. The correlation (or mutual information) oriented tools, ARACNE (39) and WGCNA (36), do not report directions of edges in networks. Bayesian-driven networks naturally reveal directed edges among the network entries. In other words, the undirected network (in left of the grey-shaded triangular) having G1, G2, and G3 entries by ARACNE and WGCNA can be differentiated into directed networks (in the right of the grey-shaded triangular), using Bayesian networks tools (48–51).
HIGH-THROUGHPUT DATA AND ITS REPOSITORIES
Currently, there are numerous types of high-throughput data (i.e., “omics”), including genomics, epigenomics, transcriptomics, metabolomics, and proteomics (15). As shown in Fig. 1B, the omics data types are aligned with the flow of genetic information in biology. Cancer genomics data in various types of cancers, including whole genome sequencing (WGS), whole exome sequencing (WES), and SNP array, has already been deposited in several public repositories including The Cancer Genome Atlas (TCGA) (11), and International Cancer Genome Consortium (ICGC) (12, 16) (Fig. 1B). Epigenomics in public databases, including the Encyclopedia of DNA Elements (ENCODE) (17) and the Database of Genotypes and Phenotypes (dbGaP) (18), possess next-generation sequencing datasets for genome-wide DNA methylation, histone modifications, transcription factor binding, and non-coding RNAs (e.g., miRNAs, piRNAs). Transcriptomic datasets are deposited in the Gene Expression Omnibus (GEO) (19), and ArrayExpress (20), for more than 10 years. Proteomics and metabolomics have now begun accumulation in the PeptieAtlas (21) and the PRoteomics IDEntifications (PRIDE) (22) databases. Each repository in Fig. 1A is not restricted to one specific data type, and users should be prudent to inspect all the data types of their interest through multiple repositories, and not a single one. The brief information of the repositories is described in Table 1.
Table 1.
Cancer-related, high-throughput data repositories. The databases in Fig. 1B are described with additional information including the number of available data sets, data types, and websites. The number of entries is deemed valid as of 05/02/2016
| Names | Description | Address | Cancer relating data |
|---|---|---|---|
| TCGA | The Cancer Genome Atlas (TCGA): now one of programs organized by newly established NCI’s Center for Cancer Genomics (11) | cancergenome.nih.gov | 34 cancer studies (types), 11,091 samples |
| dbGaP | The database of Genotypes and Phenotypes (dbGaP): archive of genome and phenotype in human | www.ncbi.nlm.nih.gov/gap | 991 datasets |
| SRA | Sequence Read Archive (SRA): raw sequencing files and alignment files from next generation sequencing | www.ncbi.nlm.nih.gov/sra | 1,950 cancer studies |
| cBioPortal | Multi-functional platform: supporting intuitive visualization, literate clinical pie chart, and simple data access (75). TCGA data visualization included. | cbioportal.org | 126 cancer genomics studies, 26,080 samples |
| ICGC | The International Cancer Genome Consortium (ICGC): global-scale cancer projects (16) | dcc.icgc.org/ | 66 cancer projects, 17,867 donors |
| ArrayExpress | An archive of functional genomics data (76) | www.ebi.ac.uk/arrayexpress | 14,974 datasets |
| EGA | The European Genome-phenome Archive (EGA) | www.ebi.ac.uk/ega/home | 1,997 datasets |
| UCSC CGB | UCSC Cancer Genomics Browser (UCSC CGB): supplying interactive heat-map based visualization, and ready-to-use tab-delimited genomics and clinical data download (77). TCGA data visualization included. | genome-cancer.ucsc.edu | 720 datasets |
| GEO | The Gene Expression Omnibus (GEO) (19): a public repository for microarray and next-generation sequencing data sets, and one of the representative repositories. | www.ncbi.nlm.nih.gov/geo | 19,554 datasets |
| ENCODE | The Encyclopedia of DNA Elements (ENCODE) Consortium: decoding functional elements in DNA (17). | www.encodeproject.org | Cancer cell lines available |
| CCLE | The Cancer Cell Line Encyclopedia (CCLE) project: genomics and visualization in about 1,000 cell lines. Drug sensitivity available for the cell lines (78). | www.broadinstitute.org/ccle/home | Genomic characterization of 1,000 cell lines |
| PeptideAtlas | An archive of proteome information (21) | www.peptideatlas.org | 99 datasets |
| PRIDE | PRoteomics IDEntifications (PRIDE) database: protein and peptide identifications, post-translational modifications (22). Mass spectrometry based proteomics data available. | www.ebi.ac.uk/pride/archive | 290 datasets |
PRIOR KNOWLEDGE
The two representative categories in prior knowledge are gene sets and pathway databases (including protein-protein interactions). A gene set consists of the relevant biological description and its gene entries. The MIT MSigDB Collections (23) (software.broadinstitute.org/gsea/msigdb/collections.jsp), one of most comprehensive repositories of gene sets, contains 13,311 entries. Recently, gene sets have begun including miRNA genes (and their expression), as well as protein-coding genes (24). By definition, however, gene sets do not contain hierarchy or mutual interaction for their gene entries (25). To accommodate such non-hierarchy, gene sets have been mainly applied to various enrichment analyses that utilize Kolmogorov–Smirnov test statistic, ANOVA, or hypergeometric test (further review in (26, 27)). A recent approach (28) identifies the conditional dependency in a gene set, to reconstruct hierarchical relationships. Thus, numerous gene sets have now been recognized as prior knowledge for use in network generation.
Unlike gene sets, pathways or protein-protein interactions have hierarchy or mutual relationships among the entries. Of numerous, diverse pathway databases, we describe the Kyoto Encyclopedia of Genes and Genomes (KEGG) (29), Reactome (30), STRING (31), and human-integrated pathway (hiPathDB) (32) databases. In particular, the KEGG (29) pathway database, one of the popular manually-curated pathway resources, consists of seven types of network contexts: cellular processes, metabolism, genetic information processing, environmental information processing, human diseases, organismal systems, and drug development (29). The KEGG pathway information is machine-readable via KGML (KEGG Markup Language). Reactome (30) is another popular peer reviewed pathway database, and contains > 6,700 reactions (e.g., phosphorylation, acetylation, etc.) extracted from 15,000 publications. For machine readability, the SBML (Systems Biology Markup Language) version of Reactome data is also available (33).
The database and web resource STRING (Search Tool for the Retrieval of Interacting Genes/Proteins, string-db.org) contains a very extensive collection of protein-protein interactions, based on publications and predictions. The interaction entries of STRING (31) amount to 932,553,897, from 2,031 organisms (as of 2016-04-19). While KEGG and Reactome both have directed network structures, STRING also has undirected network structures. The hiPathDB (32) introduces a unique concept of “superpathways,” that consolidates multiple resources of pathway databases (NCI-Nature PID (34), Reactome (30), BioCarta (35) and KEGG (29)), resulting in the most extensive hierarchical network structures.
COMPUTATIONAL MODELING AND ITS APPLICATION TO CANCER
Depending on prior knowledge usage, computational modeling, a key component in systems biology frameworks, can be divided into two modeling methods: hybrid method and data-driven method. The former incorporates prior knowledge in model development, while the latter infers networks or hypotheses directly from measurements, without prior knowledge. The tools described below are summarized in Table 2.
Table 2.
Summary of tools in network construction. The short description and homepages of some tools in the manuscript are summarized
| Class | Name | Homepage and description |
|---|---|---|
| Data-driven model | ARACNE (39) |
|
| WGCNA (36) |
|
|
| Cancer Landscapes (45) |
|
|
| Ultranet (46, 47) |
|
|
| Banjo (48) |
|
|
| CATNET (50) |
|
|
| Hybrid model | EDDY (28) |
|
| PATHOME (7) |
|
|
| SPIA (58) |
|
DATA-DRIVEN METHODS
Data-driven methods have been used to correlate mutual information as gene-gene connection for network construction (36–38), resulting in undirected networks. ARACNE (minet.meyerp.com) (39), another widely used free web-based tool, uses mutual information for constructing gene regulatory networks from transcriptome datasets. In principle, starting from all connected entries, ARACNE applies a mutual information data processing inequality (MIDPI) rule to the two adjacent edges for removing non-interacting edges (39). Since its introduction, ARACNE has been widely used in the field of cancer systems biology. Recently, ARACNE was used to describe three hypothetical stages of the epithelial-mesenchymal transition in cancer metastasis (40).
The R package, weighted gene co-expression network analysis (WGCNA, genetics.ucla.edu/Rpackages/WGCNA) (36), is another network generation tool. Specifically, WGCNA builds gene-gene co-expression networks from all pairwise correlations, among expressed genes, across the entire transcriptome. To infer connections in a network, WGCNA (36) uses a weighted adjacency matrix between gene pairs by calculating power adjacency function (41), resulting in connections among the gene entries. WGCNA has also been applied to diverse diseases, including cancer, for identifying therapeutic targets and tumorigenesis “driver” genes (42–44).
Despite the great success of correlation- and mutual information-based approaches, these approaches often generate extensive links between network entries. Consequently, methods have now been introduced to reduce non-significant links. For example, sparse inverse covariance selection (SICS) (45, 46) infers a gene regulatory network from various data types by reducing non-significant links. The main function of SCIS is to identify a subset of network entries that consists of statistically significant or optimal pairwise correlations, based on the entire correlation (equivalent to covariance) matrix between all the entries. The benefit of subset identification is that it can provide statistically direct relations with smaller number of entries. SICS methods aim at maximizing or optimizing log-likelihood of pairwise correlations, assuming pairwise correlations as Gaussian graphical models (46, 47) or multivariate Gaussian models (45). Cancer Landscapes (cancerlandscapes.org) utilizes SICS, not only to provide multiple cancer network modules, but also to integrate multilevel omics data types into statistical network modules (45).
Unlike ARACNE and WGCNA, there are several approaches to generate directed networks (Fig. 1C). Bayesian networks, another data-driven approach, utilizes a basic conditional independence (48–51). Bayesian networks is, by definition, that joint density probability of biological entries (e.g., genes) is the product of conditional probabilities of the entries in the omics data (38, 52). The definition naturally confers the ability to prune edges of the conditionally independent entries. Also, conditional dependency defines statistically casual relationships among gene entries, resulting in directed networks. The purpose of Bayesian networks is to identify the set of conditional probabilities that best describe measurements (e.g., gene expression) of biological entries in omics databases.
Banjo (users.cs.duke.edu/software) is another gene regulatory network generation tool that utilizes Bayesian network frameworks, resulting in directed networks (48). Banjo is applicable not only for single-state transcriptome data, but also for time-series data. Banjo (Bayesian network interference with Java objects) uses the multiple types of heuristic network searching to find candidate networks (equivalently, graphs), such as simulated annealing with a greedy algorithm (53), and genetic algorithm (48). The conditional probability densities of each network are estimated, and the network scores (e.g., Bayesian Information Criterion (BIC), Bayesian Dirichlet equivalence (BDe)) are then calculated. Finally, Banjo reports the network with the best score, based on its best directed edges between its entries. Banjo has also been applied to leukemia, revealing miRNA-relating network hierarchy by merging gene expression, gene regulatory networks, and copy number alterations (54).
One obstacle to all these prediction methods is that there are no “gold standards” for data-driven network generation tools. Consequently, the performance of the data-driven methods depends on data types, model parameter settings, network size, and network topology (55).
HYBRID METHODS
In hybrid methods, models are generated to analyze high-throughput data via prior knowledge (e.g., gene sets, pathways) (56), resulting in network inference. Traditionally to date, hybrid methods use pathways as prior knowledge. Recently, gene sets have been recognized as another prior knowledge source for inferring networks that consist of entries and their mutual interactions.
Another tool, EDDY (evaluation of dependency differentiality) (28) considers two conditions, and applies Bayesian networks framework to all the gene sets. EDDY selects the best network structure for each gene set, by using Jensen-Shannon (JS) divergences and permutation tests from all possible network structures. The tool then calculates the two probability density distributions of a network structure for the two conditions. Subsequently, EDDY calculates JS divergence for the two distributions of the network structure, measuring JS divergence as the difference of the two distributions. The significance of JS divergence is measured by the permutation test, identifying the best network structure having statistically significant JS divergence. The output is a network that consists of the entries (of the gene set) and their interactions between the entries. The tool was recently applied to glioblastoma multiforme (GBM), resulting in the successful identification of specific molecular subtypes of glioblastoma (28).
Prior pathway information with omics data has been incorporated into statistical frameworks for the past ten years (7, 8, 57), successfully generating network structures. In this approach, the challenge to build the statistical framework is developing and defining a statistic reflecting pathway topology. Pathway topology indicates interaction types (e.g., activation, inhibition, modification) as well as order (e.g., upstream, downstream) of biological entries. Another tool, SPIA (signaling pathway impact analysis) (58) (bioconductor.org/packages/release/bioc/html/SPIA.html), utilizes the KEGG pathway database as prior knowledge. Instead of utilizing the individual signaling molecules (in KEGG pathways), SPIA aligns the consecutive KEGG signaling “flows” with omics data. Additionally, SPIA now considers two types of a flow between two adjacent signaling molecules: activation and inhibition. SPIA quantitatively measures influence (i.e., perturbation statistic in a given pathway) on signal cascading flows by using omics data between two experimental groups. For any given pathway, SPIA obtains P values for the perturbation statistic by using permutation tests. SPIA also reconstructs statistically significant pathways in a network. Recently, SPIA was applied to aggressive prostate cancer, discovering that the disease shares a pathway network with small cell lung cancer (59).
We also developed pathway topology-driven hybrid methods (7, 8), specifically for network generation, including PATHOME (7). These two methods also input the KEGG database (29) as prior knowledge for network generation. The earlier algorithm (8) (henceforth, pre-PATHOME) identified subsets of all KEGG pathways by utilizing permutation-oriented statistical tests, based on a whole transcriptome. Since graphical structures of the KEGG pathways are too complex, we decomposed to all the possible paths (~130 million, equivalently, subpathways) by traversing the graph structures.
In pre-PATHOME, each path consists of biological entries and their mutual interactions between adjacent two entries, either activation or inhibition. Given a subpathway, we devised a statistic to consider interactions (equivalently, edges) of two adjacent entries, as well as orders of biological entities (8). We assumed the first order Markov property (denoted as Fedge in (8)) where the fold-changes of the entities were regarded as observations. Subsequently, we performed permutation-based statistical tests for the product of Fedge and two additional statistics in each path. The statistically significant paths were collected and visualized. The pre-PATHOME was applied to an early onset colorectal cancer (CRC) dataset (60), revealing the pathways of epithelial-to-mesenchymal transition and immunosuppression even in normal adjacent cells of the CRC patients (8). The pre-PATHOME (8) was also deployed to identify trastuzumab-resistance pathways relating to networks in HER2(+) breast cancer (61), revealing five biomarker candidates associated with trastuzumab non-responsiveness (ATF4, CHEK2, ENAH, ICOSLG, and RAD51).
Our group recently developed another hybrid method, PATHOME, (7). The pre-PATHOME (8) assumed that all interactions in a subpathway are dependent on their upstream entities (the so called, first order Markov property). PATHOME assumes that all edges in a subpathway are independent, adopting a two-stage strategy in our statistical framework (7). In the first stage, out of 130 million KEGG subpathways, PATHOME selects those with their edges aligned with correlations. In the second stage, we test the selected subpathways under the null hypothesis, that no differential correlation patterns between two groups are observed. Despite the independence assumption among edges, PATHOME showed better agreement with a cancer signaling reference set (62), when compared to other gene set analysis tools (e.g., DAVID (63), and GSEA (25)).
PATHOME has also been applied for delineating druggable target candidates, as well as molecular mechanisms, in both gastric and breast cancers (7, 64, 65). Recently, PATHOME was applied to gastric cancer (GC) transcriptome datasets, suggesting a HNF4α/WNT5A axis to be a new druggable signaling, as well as having a clinical relevance in diffuse type GC (64, 65). Since trastuzumab treatment of HER2-positive GC tumors show limited benefit, compared with ERBB2-positive breast cancer (66), PATHOME was applied to high ERBB2 (equivalently, HER2)-expressing GC patient datasets in the TCGA (64, 67). In these analyses, PATHOME revealed that NFBIE, PTK2, and PIK3CA, all downstream molecules of ERBB2, associate with genomic characteristics of high ERBB2-expressing GC patients over low ERBB2-expressing GC patients (64).
CONCLUSIONS
Systems biology is a general modeling framework that utilizes high-throughput data and prior knowledge, to result in network inference and hypotheses suggestions. Most network generation tools are based on whole transcriptome data. Using statistical models, the integration of other data types into network topology is still challenging. For example, for effective targeted therapy, the effects of mutations need to be incorporated into pathway topology under the systems biology frameworks (68). Also, for facilitation of translating cancer big data toward therapeutic benefit, pharmacokinetics/pharmacodynamics assessments (69–71) need to be considered in network generation in future.
Although this review does not describe visualization tools, intuitive and informative graphical visualization of the models should keep pace with systems biology tools (72–74).
ACKNOWLEDGEMENTS
This work was supported by the Gachon University Gil Medical Center (Grant number: 2016-06), and performed by a subproject of KISTI (Korea Institute of Science and Technology Information)’s project No. P16018 (Development of HPC-based Big Data for healthy Aging Society) funded by (Ministry of Science, ICT, and Future Planning). The Author thanks Curt Balch for editing the manuscript.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Werner HM, Mills GB, Ram PT. Cancer Systems Biology: a peek into the future of patient care? Nat Rev Clin Oncol. 2014;11:167–176. doi: 10.1038/nrclinonc.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soon WW, Hariharan M, Snyder MP. High-throughput sequencing for biology and medicine. Mol Syst Biol. 2013;9:640. doi: 10.1038/msb.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chuang HY, Hofree M, Ideker T. A decade of systems biology. Annu Rev Cell Dev Biol. 2010;26:721–744. doi: 10.1146/annurev-cellbio-100109-104122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jost D, Nowojewski A, Levine E. Small RNA biology is systems biology. BMB Rep. 2011;44:11–21. doi: 10.5483/BMBRep.2011.44.1.11. [DOI] [PubMed] [Google Scholar]
- 5.Nam S, Long X, Kwon C, Kim S, Nephew KP. An integrative analysis of cellular contexts, miRNAs and mRNAs reveals network clusters associated with antiestrogen-resistant breast cancer cells. BMC Genomics. 2012;13:732. doi: 10.1186/1471-2164-13-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rho S, You S, Kim Y, Hwang D. From proteomics toward systems biology: integration of different types of proteomics data into network models. BMB Rep. 2008;41:184–193. doi: 10.5483/BMBRep.2008.41.3.184. [DOI] [PubMed] [Google Scholar]
- 7.Nam S, Chang HR, Kim KT, et al. PATHOME: an algorithm for accurately detecting differentially expressed subpathways. Oncogene. 2014;33:4941–4951. doi: 10.1038/onc.2014.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nam S, Park T. Pathway-based evaluation in early onset colorectal cancer suggests focal adhesion and immunosuppression along with epithelial-mesenchymal transition. PLoS One. 2012;7:e31685. doi: 10.1371/journal.pone.0031685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altaf-Ul-Amin M, Afendi FM, Kiboi SK, Kanaya S. Systems biology in the context of big data and networks. Biomed Res Int. 2014;2014;428570 doi: 10.1155/2014/428570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marx V. Drilling into big cancer-genome data. Nat Meth. 2013;10:293–297. doi: 10.1038/nmeth.2410. [DOI] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Baran J, Cros A, et al. International Cancer Genome Consortium Data Portal--a one-stop shop for cancer genomics data. Database (Oxford) 20112011:bar026. doi: 10.1093/database/bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh S, Matsuoka Y, Asai Y, Hsin KY, Kitano H. Software for systems biology: from tools to integrated platforms. Nat Rev Genet. 2011;12:821–832. doi: 10.1038/nrg3096. [DOI] [PubMed] [Google Scholar]
- 14.Zierer J, Menni C, Kastenmuller G, Spector TD. Integration of ‘omics’ data in aging research: from biomarkers to systems biology. Aging Cell. 2015;14:933–944. doi: 10.1111/acel.12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pecina-Slaus N, Pecina M. Only one health, and so many omics. Cancer Cell Int. 2015;15:64. doi: 10.1186/s12935-015-0212-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Cancer Genome Consortium. Hudson TJ, Anderson W, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tryka KA, Hao L, Sturcke A, et al. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rocca-Serra P, Brazma A, Parkinson H, et al. ArrayExpress: a public database of gene expression data at EBI. C R Biol. 2003;326:1075–1078. doi: 10.1016/j.crvi.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 21.Kusebauch U, Deutsch EW, Campbell DS, Sun Z, Farrah T, Moritz RL. Using PeptideAtlas, SRMAtlas, and PASSEL: Comprehensive Resources for Discovery and Targeted Proteomics. Curr Protoc Bioinformatics. 2014;46:13.25.1–13.25.28. doi: 10.1002/0471250953.bi1325s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones P, Cote R. The PRIDE proteomics identifications database: data submission, query, and dataset comparison. Methods Mol Biol. 2008;484:287–303. doi: 10.1007/978-1-59745-398-1_19. [DOI] [PubMed] [Google Scholar]
- 23.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res. 2009;37:W356–362. doi: 10.1093/nar/gkp294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maciejewski H. Gene set analysis methods: statistical models and methodological differences. Brief Bioinform. 2014;15:504–518. doi: 10.1093/bib/bbt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emmert-Streib F, Tripathi S, de Matos Simoes R. Harnessing the complexity of gene expression data from cancer: from single gene to structural pathway methods. Biol Direct. 2012;7:44. doi: 10.1186/1745-6150-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung S, Kim S. EDDY: a novel statistical gene set test method to detect differential genetic dependencies. Nucleic Acids Res. 2014;42:e60. doi: 10.1093/nar/gku099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croft D, Mundo AF, Haw R, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu N, Seo J, Rho K, et al. hiPathDB: a human-integrated pathway database with facile visualization. Nucleic Acids Res. 2012;40:D797–802. doi: 10.1093/nar/gkr1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hucka M, Finney A, Sauro HM, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 34.Schaefer CF, Anthony K, Krupa S, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimura D. BioCarta. Biotech Software & Internet Report. 2001;2:117–120. doi: 10.1089/152791601750294344. [DOI] [Google Scholar]
- 36.Allen JD, Xie Y, Chen M, Girard L, Xiao G. Comparing statistical methods for constructing large scale gene networks. PLoS One. 2012;7:e29348. doi: 10.1371/journal.pone.0029348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butte AJ, Kohane IS. Mutual information relevance networks: functional genomic clustering using pairwise entropy measurements. Pac Symp Biocomput. 2000:418–429. doi: 10.1142/9789814447331_0040. [DOI] [PubMed] [Google Scholar]
- 38.Markowetz F, Spang R. Inferring cellular networks--a review. BMC Bioinformatics. 2007;8(Suppl 6):S5. doi: 10.1186/1471-2105-8-S6-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Margolin AA, Nemenman I, Basso K, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka H, Ogishima S. Network biology approach to epithelial-mesenchymal transition in cancer metastasis: three stage theory. J Mol Cell Biol. 2015;7:253–266. doi: 10.1093/jmcb/mjv035. [DOI] [PubMed] [Google Scholar]
- 41.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:12287. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 42.Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 43.Gnad F, Doll S, Manning G, Arnott D, Zhang Z. Bioinformatics analysis of thousands of TCGA tumors to determine the involvement of epigenetic regulators in human cancer. BMC Genomics. 2015;16(Suppl 8):S5. doi: 10.1186/1471-2164-16-S8-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horvath S, Zhang B, Carlson M, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kling T, Johansson P, Sanchez J, Marinescu VD, Jornsten R, Nelander S. Efficient exploration of pan-cancer networks by generalized covariance selection and interactive web content. Nucleic Acids Res. 2015;43:e98. doi: 10.1093/nar/gkv413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jarvstrat L, Johansson M, Gullberg U, Nilsson B. Ultranet: efficient solver for the sparse inverse covariance selection problem in gene network modeling. Bioinformatics. 2013;29:511–512. doi: 10.1093/bioinformatics/bts717. [DOI] [PubMed] [Google Scholar]
- 47.Storry JR, Joud M, Christophersen MK, et al. Homozygosity for a null allele of SMIM1 defines the Vel-negative blood group phenotype. Nat Genet. 2013;45:537–541. doi: 10.1038/ng.2600. [DOI] [PubMed] [Google Scholar]
- 48.Yu J, Smith VA, Wang PP, Hartemink AJ, Jarvis ED. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics. 2004;20:3594–3603. doi: 10.1093/bioinformatics/bth448. [DOI] [PubMed] [Google Scholar]
- 49.Frolova A, Wilczyński B. Distributed Bayesian Networks Reconstruction on the Whole Genome Scale. bioRxiv. 2015 doi: 10.1101/016683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salzman P, Almudevar A. Using complexity for the estimation of Bayesian networks. Stat Appl Genet Mol Biol. 2006;5 doi: 10.2202/1544-6115.1208. Article21. [DOI] [PubMed] [Google Scholar]
- 51.Chen X, Chen M, Ning K. BNArray: an R package for constructing gene regulatory networks from microarray data by using Bayesian network. Bioinformatics. 2006;22:2952–2954. doi: 10.1093/bioinformatics/btl491. [DOI] [PubMed] [Google Scholar]
- 52.Bansal M, Belcastro V, Ambesi-Impiombato A, di Bernardo D. How to infer gene networks from expression profiles. Mol Syst Biol. 2007;3:78. doi: 10.1038/msb4100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adabor ES, Acquaah-Mensah GK, Oduro FT. SAGA: a hybrid search algorithm for Bayesian Network structure learning of transcriptional regulatory networks. J Biomed Inform. 2015;53:27–35. doi: 10.1016/j.jbi.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 54.Volinia S, Galasso M, Costinean S, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madhamshettiwar PB, Maetschke SR, Davis MJ, Reverter A, Ragan MA. Gene regulatory network inference: evaluation and application to ovarian cancer allows the prioritization of drug targets. Genome Med. 2012;4:41. doi: 10.1186/gm340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galvanauskas V, Simutis R, Lubbert A. Hybrid process models for process optimisation, monitoring and control. Bioprocess Biosyst Eng. 2004;26:393–400. doi: 10.1007/s00449-004-0385-x. [DOI] [PubMed] [Google Scholar]
- 57.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8:e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tarca AL, Draghici S, Khatri P, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25:75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith BA, Sokolov A, Uzunangelov V, et al. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci U S A. 2015;112:E6544–6552. doi: 10.1073/pnas.1518007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hong Y, Ho KS, Eu KW, Cheah PY. A susceptibility gene set for early onset colorectal cancer that integrates diverse signaling pathways: implication for tumorigenesis. Clin Cancer Res. 2007;13:1107–1114. doi: 10.1158/1078-0432.CCR-06-1633. [DOI] [PubMed] [Google Scholar]
- 61.Nam S, Chang HR, Jung HR, et al. A pathway-based approach for identifying biomarkers of tumor progression to trastuzumab-resistant breast cancer. Cancer Lett. 2015;356:880–890. doi: 10.1016/j.canlet.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 62.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 63.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 64.Chang HR, Nam S, Kook MC, et al. HNF4alpha is a therapeutic target that links AMPK to WNT signalling in early-stage gastric cancer. Gut. 2016;65:19–32. doi: 10.1136/gutjnl-2014-307918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang HR, Park HS, Ahn YZ, et al. Improving gastric cancer preclinical studies using diverse in vitro and in vivo model systems. BMC Cancer. 2016;16:200. doi: 10.1186/s12885-016-2232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 67.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hernansaiz-Ballesteros RD, Salavert F, Sebastian-Leon P, Aleman A, Medina I, Dopazo J. Assessing the impact of mutations found in next generation sequencing data over human signaling pathways. Nucleic Acids Res. 2015;43:W270–275. doi: 10.1093/nar/gkv349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Griffith M, Griffith OL, Coffman AC, et al. DGIdb: mining the druggable genome. Nat Methods. 2013;10:1209–1210. doi: 10.1038/nmeth.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wishart DS, Knox C, Guo AC, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Franz M, Lopes CT, Huck G, Dong Y, Sumer O, Bader GD. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2016;32:309–311. doi: 10.1093/bioinformatics/btv557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jang Y, Yu N, Seo J, Kim S, Lee S. MONGKIE: an integrated tool for network analysis and visualization for multi-omics data. Biol Direct. 2016;11:10. doi: 10.1186/s13062-016-0112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parkinson H, Sarkans U, Kolesnikov N, et al. ArrayExpress update--an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39:D1002–1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu J, Sanborn JZ, Benz S, et al. The UCSC Cancer Genomics Browser. Nat Methods. 2009;6:239–240. doi: 10.1038/nmeth0409-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]