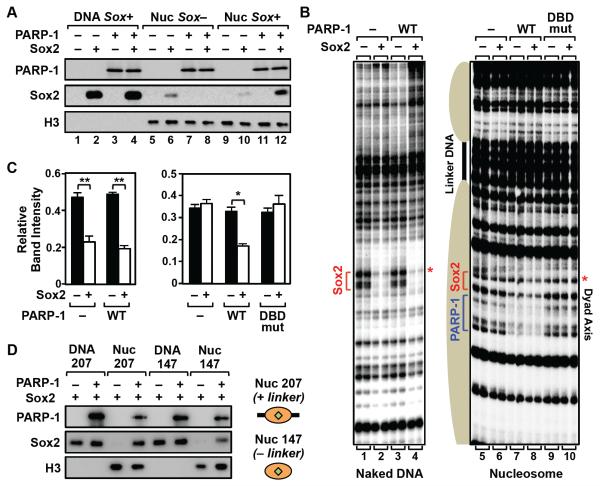
Figure 5. PARP-1 stabilizes Sox2 binding to nucleosome in vitro.
A) In vitro nucleosome binding assays show PARP-1-dependent binding of Sox2 to nucleosomes, but not naked DNA. Ten nM of biotin-labeled 601 NPE DNA (207 bp) with or without a Sox motif (Sox+ or Sox−, respectively), or the same DNA assembled into a mononucleosome (Nuc), was immobilized on streptavidin beads and incubated ± recombinant Sox2 (10 nM) and PARP-1 (10 nM), as indicated. After washing, the bound proteins were analyzed by Western blotting.
B) In vitro DNase I footprinting assays show PARP-1-dependent binding of Sox2 to nucleosomes, but not naked DNA. Left, Footprinting assay with naked 3x 601 NPE DNA containing a Sox motif. Right, Footprinting assay with a trinucleosome containing a Sox motif located at the dyad axis of the middle nucleosome. Addition of 10 nM WT PARP-1, but not a DNA binding domain mutant PARP-1 (DBDmut), enhances Sox2 (25 nM) binding, as indicated. PARP-1 binding to the nucleosome is also evident.
C) Quantification of the DNase I footprinting assays shown in panel C. The bands marked with a red asterisk in panel B were quantified using a phosphorimager and ImageJ software. Each bar represents the mean plus the SEM, n = 3. Asterisks indicate significant differences versus the control (−Sox2) (Student’s t test, ** p < 0.01 and * p < 0.05).
D) PARP-1 stabilizes Sox2 binding to nucleosomes with or without linker DNA. In vitro nucleosome binding assay, as in panel A, using 601 NPE mononucleosomes with (Nuc 207; 207 bp) or without (Nuc 147; 147 bp) linker DNA, or the corresponding naked DNA (“DNA”).