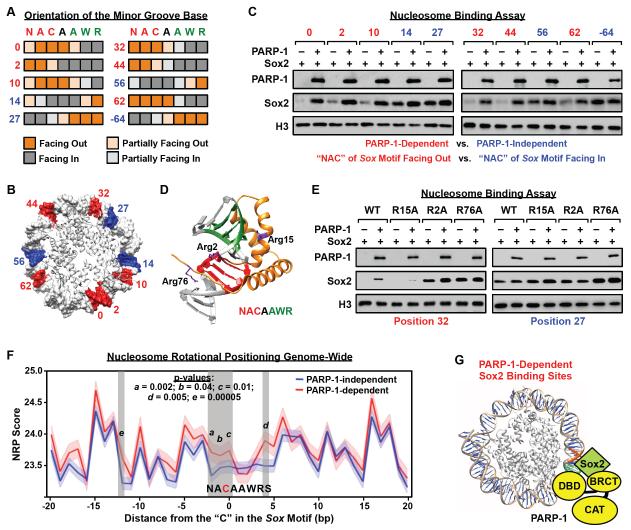
Figure 7. Cooperative interactions with PARP-1 facilitate Sox2 binding to suboptimal sites in nucleosomes.
A) Orientation of the minor groove base relative to the histone octamer for each base pair in the Sox motif in the 601 NPE nucleosome, as determined from high resolution X-ray crystal structures (PBD: 3LZ0, 3LZ1, 3MVD). Heatmaps showing the orientation of each position in the Sox motif (orange shading, minor groove facing away from the histone octamer; gray shading, minor groove facing toward the histone octamer). Letters indicate the nucleotides at each position of the Sox motif (N = A, C, G, or T; W = A or T; R = A or G) and numbers indicate the position of the Sox motif on the nucleosome, as shown in panel B.
B) Position of the “NAC” in the Sox motif mapped on the 601 NPE nucleosome for a series of different constructs where the Sox motif has been moved along the length of the DNA. Red, PARP-1-dependent Sox2 binding site. Blue: PARP-1-independent Sox2 binding site. Only half of the nucleosome is shown. The numbering is based on the distance of the “C” in the first half of the Sox motif to the dyad axis of the nucleosome (the −64 position is not shown).
C) The rotational orientation of the Sox motif determines the PARP-1-dependence of Sox2 binding to nucleosomes. In vitro nucleosome binding assays, with biotin-labeled 601 NPE DNA (207 bp) as in Fig 5A, using a set of nucleosome templates with a Sox motif placed in different translational positions across the nucleosome, as shown in panel B.
D) X-ray crystal structure of Sox2 HMG domain binding to DNA (1GT0) (Remenyi et al., 2003) with the locations of three key arginine residues (Arg2, Arg15, Arg76) indicated (side chains shown in purple).
E) Arginine residues in the HMG domain mediate PARP-1-dependent binding of Sox2 to nucleosomes. In vitro nucleosome binding assays, as in panel C, using a set of Sox2 Arg → Ala mutant proteins (R2A, R15A, R76A). The Sox2 mutants were assayed with two different nucleosome constructs: “position 32” (PARP-1-dependent) and “position 27” (PARP-1-independent).
F) NRP scores associated with PARP-1-dependent and PARP-1-independent Sox2 binding sites in mESCs. See Fig. S7H for an outline of the methods. The x-axis represents the position in nucleotides of the Sox motif associated with the Sox2 binding site, as determined by ChIP-seq, with the “C” in the first half of the Sox motif set to 0. The y-axis represents average NRP score associated with each nucleotide position (dark lines), with the SEM range indicated by lighter shading. Vertical grey shading highlights regions exhibiting a significant difference in NRP score between PARP-1-dependent and PARP-1-independent Sox2 binding sites. Higher NRP scores indicate a greater tendency for the sequence to face away from the histone octamer (see Fig. S7B).
G) Model for Sox2 binding to nucleosomes at PARP-1-dependent sites. PARP-1 interacts with nucleosomal DNA through its DNA binding domain (DBD) and Sox2 through its BRCT motif. PARP-1 stabilizes Sox2 interaction with nucleosomal DNA when the Sox motif sequence is positioned unfavorably along the nucleosome for Sox2 to bind.
See also Fig. S7.