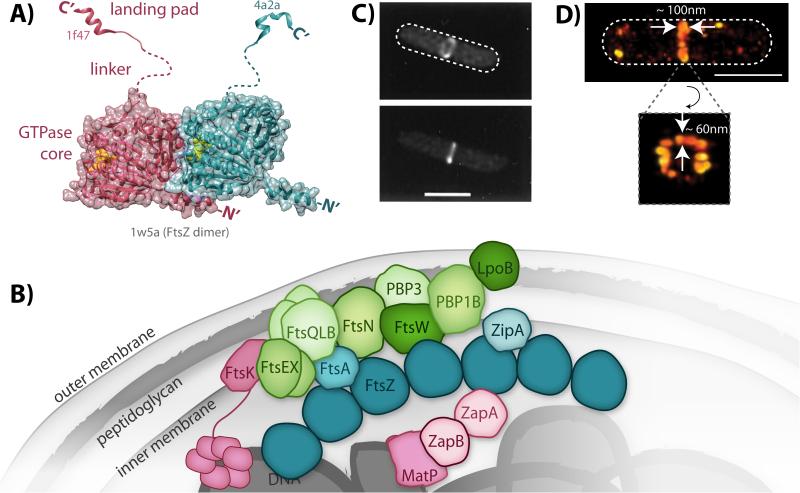
Figure 1.
Structure and organization of FtsZ and the divisome. A: Model of a FtsZ dimer based on solved crystal structures. The two FtsZ subunits within the dimer are colored individually in either magenta or teal. Bound GTP (yellow spheres) is observed at the polymerization interface in the dimer structure of the globular GTPase core domains (PDB ID: 1w5a). The two different landing pad structures were solved in complex with either ZipA (magenta, PDB ID: 1f47) or FtsA (teal, PDB ID: 4a2a). The unstructured linker domains are represented with dashed lines. B: Schematic of the network of interacting divisome proteins involved in cytokinesis. For simplicity, only FtsZ (dark teal) is shown as an oligomer although other divisome constituents may also oligomerize. Divisome constituents are broadly grouped as the Z-ring (FtsZ and its membrane tethers, teal), nucleoid-associated (magenta), and peptidoglycan-associated (green), although some proteins span multiple categories. C: Early images of the Z-ring obtained by conventional fluorescence microscopy of E. coli cells expressing FtsZ-GFP [108]. (© 1996 National Academy of Sciences) D: Recent superresolution images of Z-ring heterogeneity in E. coli obtained by photoactivated localization microscopy [21]. White dashed lines depict approximate cell outlines. Scale bars, 1 μm.