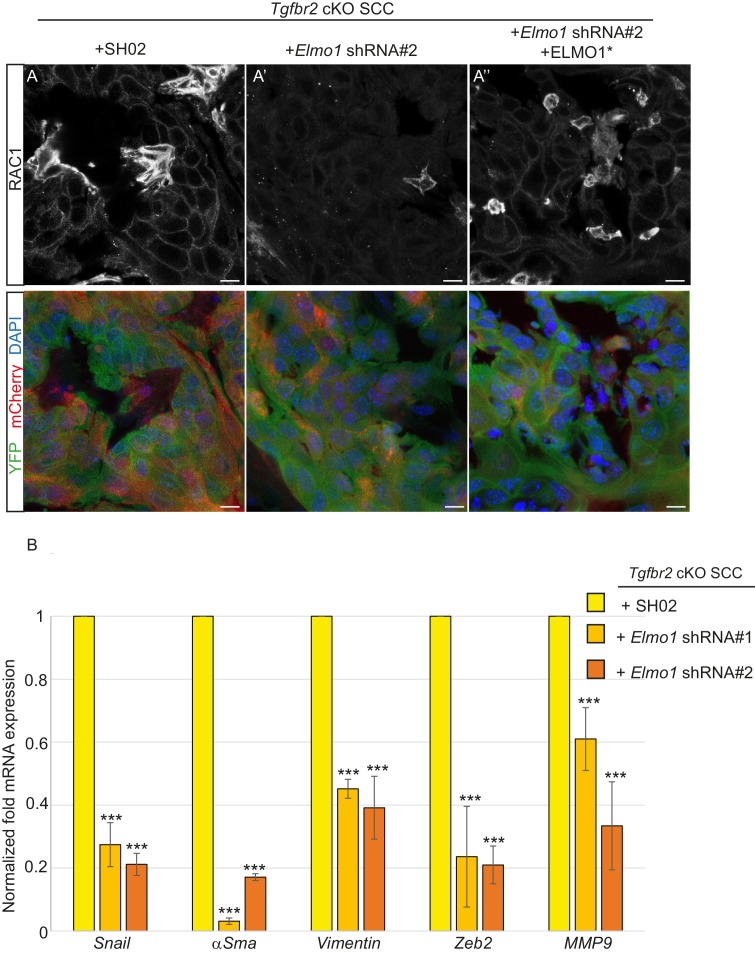
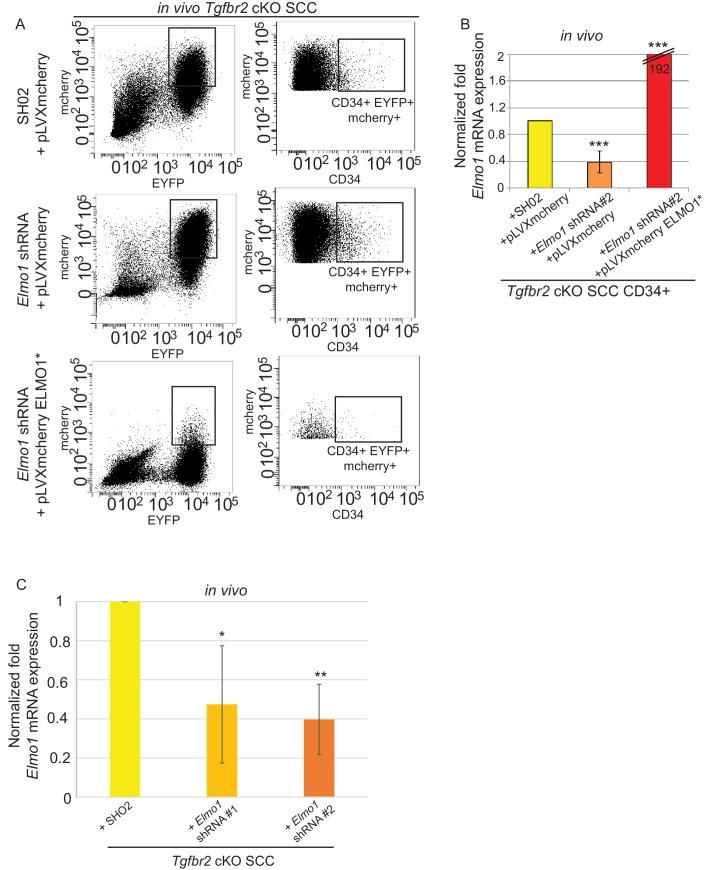
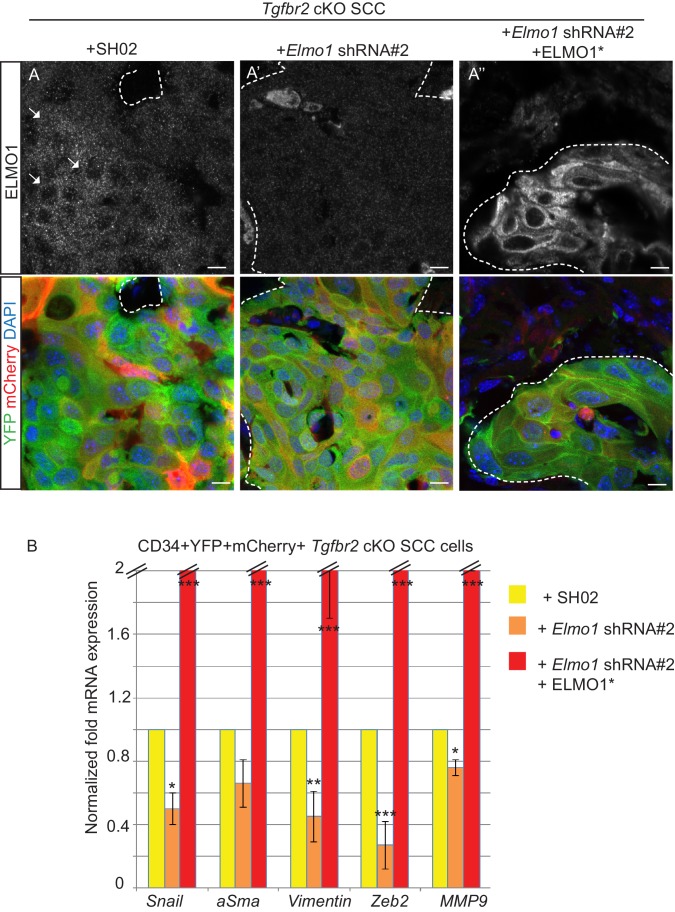
Figure 9. Knockdown of Elmo1 diminishes RAC1 expression and markers of invasion in Tgfbr2-deficient SCC.
(A) Immunofluorescence staining with antibodies against YFP and RAC1 revealed a reduction in RAC1 staining at the tumor-stroma border in Tgfbr2 cKO tumors with knockdown of Elmo1 (A’), compared to SH02 control tumors (A). Tumors expressing the hairpin-resistant ELMO1* construct show restoration of RAC1 compared to tumors with Elmo1 knockdown (A’’). All pictures have been taken at the same exposure time in the RAC1 channel. Three tumors for each condition have been analyzed. DAPI counterstains all nuclei in blue. Scale bars = 10 μm. (B) qPCR analysis of genes implicated in EMT, invasion and metastasis revealed a significant reduction in their mRNA expression in CD34+ cells isolated from Tgfbr2 cKO tumors with knockdown of Elmo1 compared to SH02 control tumors (see Figure 9—figure supplement 1 and 2). Data represent the mean ± standard deviation. Asterisks denote statistical significance using two way-ANOVA and Bonferroni post tests to compare each Elmo1 shRNA to SHO2 control. All p-values are <0.001.
DOI: http://dx.doi.org/10.7554/eLife.22914.027