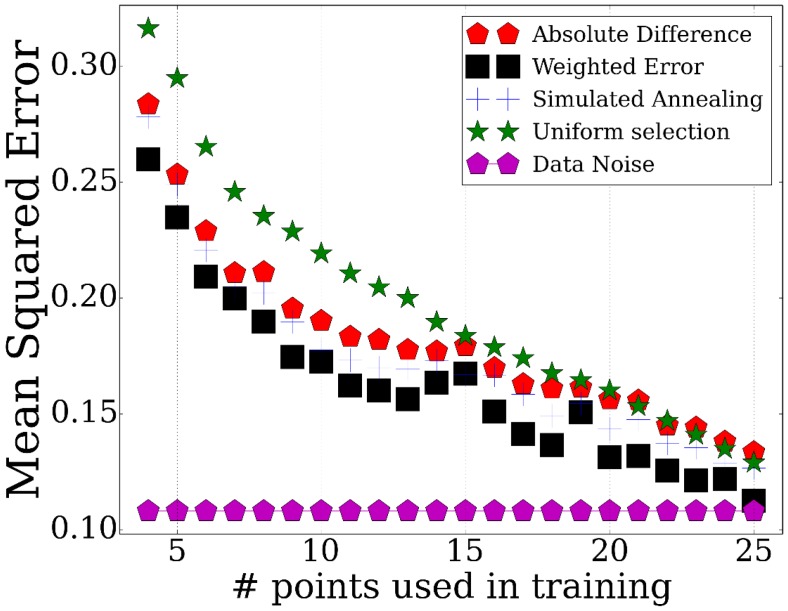
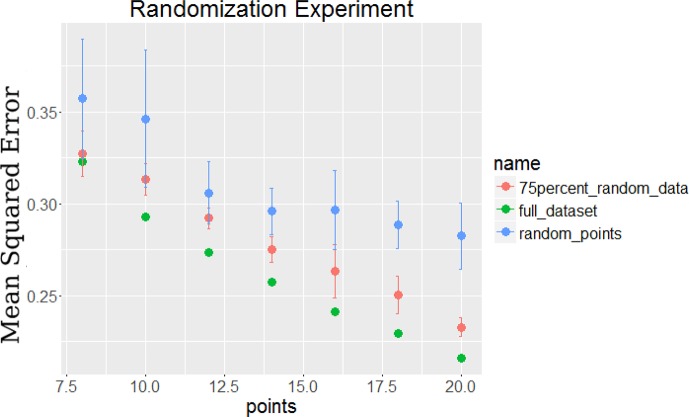
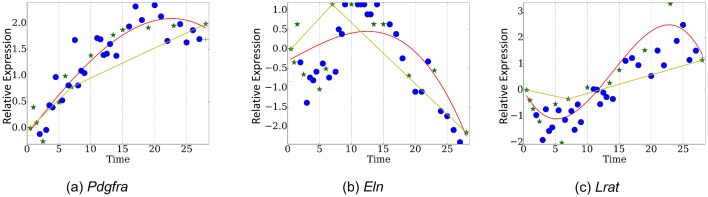
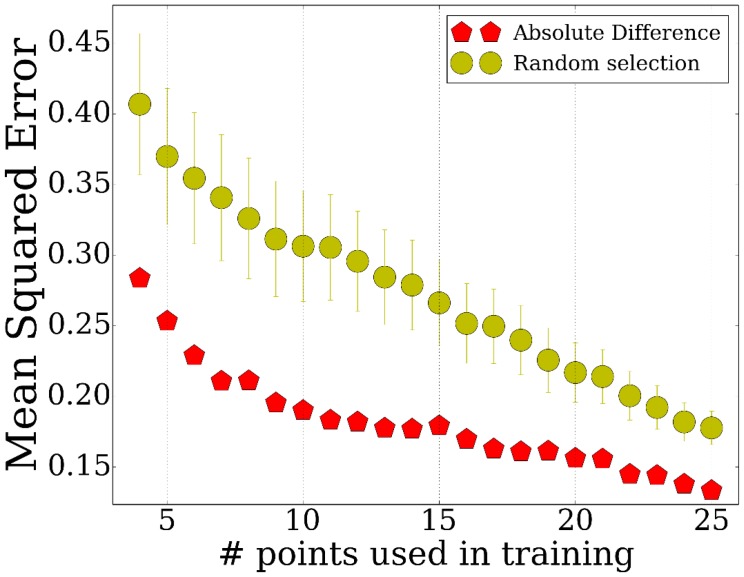
Figure 2. Performance of TPS using different sizes for the selected points.
Error comparisons of TPS variants to uniform selection of points and noise. Absolute difference - Greedy iterative addition with absolute difference initialization (Algorithm 1, Appendix Methods). Simulated annealing - Iterating using simulated annealing with absolute difference initialization. Weighted error - Selection based on cluster rather than individual gene errors. See Appendix Methods for details.
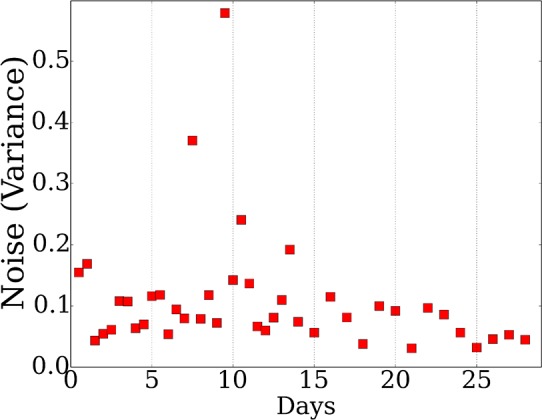
Figure 2—figure supplement 1. Average noise in each mRNA expression time point.