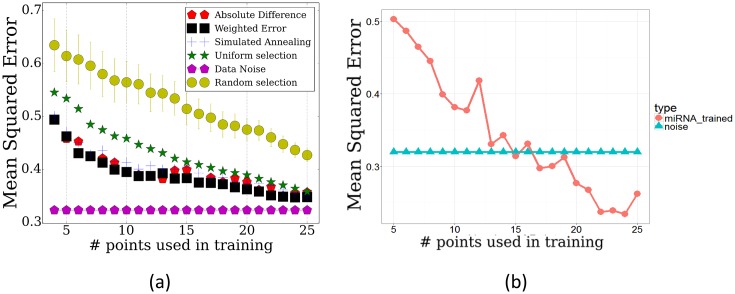
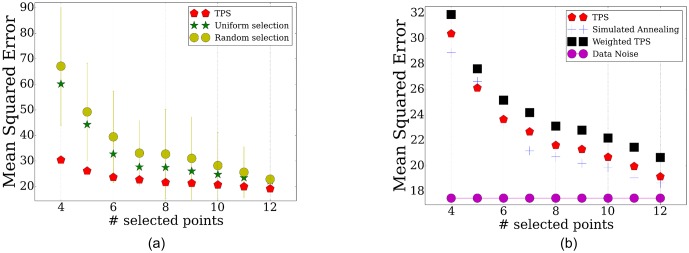
Figure 4. Performance of TPS by on the miRNA data.
(a) TPS reconstruction error when using the mRNA data to select time points for the miRNA experiments. Results of random and uniform selection as well as repeat noise error are also presented for comparison. TPS variants shown are the same two presented in Figure 2. (b) Error of splines with points selected by training TPS on the actual miRNA data itself, using the maximum absolute difference initialization.
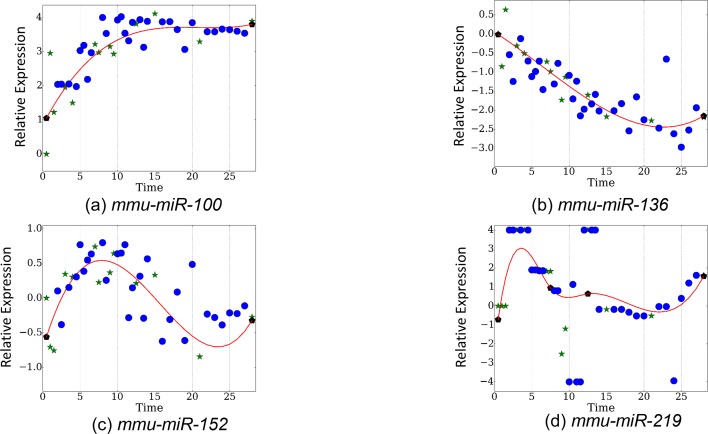
Figure 4—figure supplement 1. Observed and reconstructed expression proles for miRNAs (a) mmu-miR-100, (b) mmu-miR-136,c) mmu-miR-152, (d) mmu-miR-219.
Figure 4—figure supplement 2. 8 stable miRNA clusters.