Figure 2. Genomic Characterization of NKX2-1 Binding in the MGE.
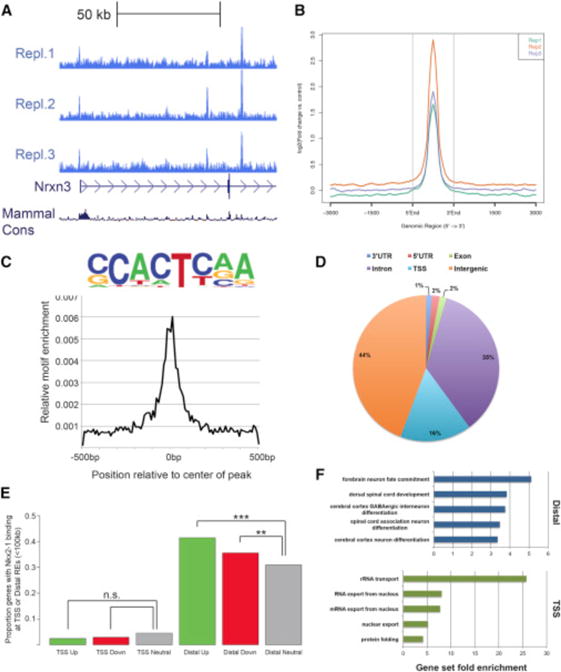
(A) Biological replicates of NKX2-1 ChIP-seq showing consistent NKX2-1 binding at the 5′ end of Nrxn3.
(B) Average profile of NKX2-1 binding at called ChIP-seq peaks comparing the three biological replicates.
(C) NKX2-1 consensus motif derived from de novo motif analysis of NKX2-1 ChIP-seq data.
(D) Genomic features of NKX2-1-bound REs.
(E) Proportion of differentially expressed genes in the Nkx2-1cKO with TSS and distal NKX2-1 binding. Fisher’s exact test was used to test significance between the groups: ***p < 0.001, **p < 0.01.
(F) Enriched gene ontology annotations of putative NKX2-1 binding target genes for distal REs (blue) and NKX2-1-bound TSSs (green).
