Figure 6. NKX2-1 and LHX6 Co-occupy aREs in the MGE.
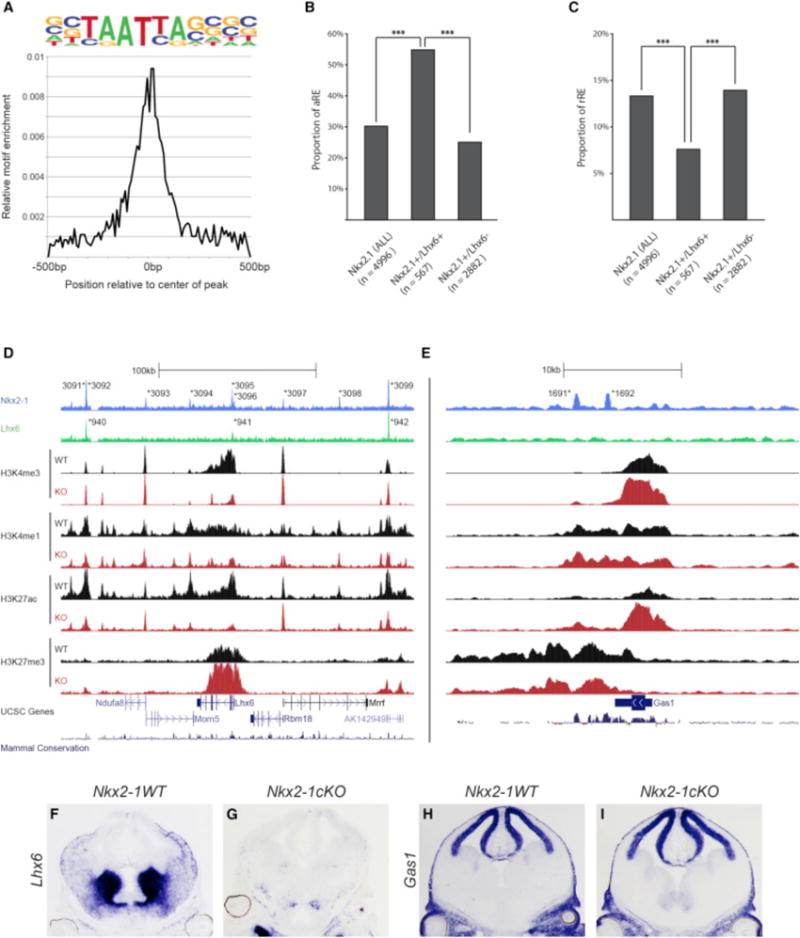
(A) Enrichment of primary LHX6 de novo motif in relation to the center of LHX6-bound REs.
(B) Proportion of aREs in NKX2-1+ (all), NKX2-1+/LHX6+, and Nkx2-1+/Lhx6− bound REs. Chi-square test was used to test significance between the groups: ***p < 0.001.
(C) Proportion of rREs in NKX2-1+ (all), NKX2-1+/LHX6+, and Nkx2-1+/Lhx6− bound REs. Fisher’s exact test was used to test significance between the groups: ***p < 0.001.
(D and E) Genomic region of the Lhx6 (D) and Gas1 (E) loci (+/−100 kb from TSS); NKX2-1 ChIP-seq, LHX6 ChIP-seq, H3K4me1, H3K4me3, H3K27Ac, H3K27me3, UCSC genes, and mammalian conservation. Both WT and Nkx2-1cKO data are shown for the H3 modifications. Called peaks are labeled with an asterisk.
(F–I) In situ analysis of Lhx6 (F and G) and Gas1 (H and I) transcription in WT and Nkx2-1cKO forebrain at E13.5.
